Abstract
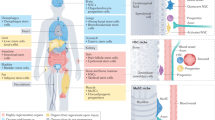
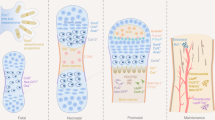
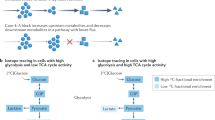
Over the past decade, it has become increasingly clear that many tissues have regenerative capabilities. The challenge has been to find the stem cells or progenitors that are responsible for tissue renewal and repair. The revolution in technological advances that permit sophisticated spatial, temporal and kinetic analyses of stem cells has allowed stem cell hunters to ferret out where stem cells live, and to monitor when they come and go from these hiding places.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Schofield, R. The relationship between the spleen colony-forming cell and the haematopoietic stem cell. Blood Cells 4, 7–25 (1978).
Till, J. E. & McCulloch, E. A. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 14, 213–222 (1961).
Punzel, M. & Ho, A. D. Divisional history and pluripotency of human hematopoietic stem cells. Ann. N. Y. Acad. Sci. 938, 72–81; discussion 81–82 (2001).
Potten, C. S. Keratinocyte stem cells, label-retaining cells and possible genome protection mechanisms. J. Investig. Dermatol. Symp. Proc. 9, 183–195 (2004).
Fuchs, E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell 137, 811–819 (2009).
Cotsarelis, G., Sun, T. T. & Lavker, R. M. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61, 1329–1337 (1990).
Cotsarelis, G., Cheng, S. Z., Dong, G., Sun, T. T. & Lavker, R. M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell 57, 201–209 (1989).
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004).
Blanpain, C., Lowry, W. E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004).
Claudinot, S., Nicolas, M., Oshima, H., Rochat, A. & Barrandon, Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc. Natl Acad. Sci. USA 102, 14677–14682 (2005).
Waghmare, S. K et al. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 27, 1309–1320 (2008).
Hsu, Y. C., Pasolli, H. A. & Fuchs, E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105 (2011).
Nowak, J. A., Polak, L., Pasolli, H. A. & Fuchs, E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3, 33–43 (2008).
Spangrude, G. J., Heimfeld, S. & Weissman, I. L. Purification and characterization of mouse hematopoietic stem cells. Science 241, 58–62 (1988).
Cheshier, S. H., Morrison, S. J., Liao, X. & Weissman, I. L. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc. Natl Acad. Sci. USA 96, 3120–3125 (1999).
Kiel, M. J. et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature 449, 238–242 (2007).
Jetmore, A. et al. Homing efficiency, cell cycle kinetics, and survival of quiescent and cycling human CD34(+) cells transplanted into conditioned NOD/SCID recipients. Blood 99, 1585–1593 (2002).
Zhang, J. et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836–841 (2003).
Foudi, A. et al. Analysis of histone 2B–GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27, 84–90 (2009).
Wilson, A. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008).
Ema, H. et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat. Protoc. 1, 2979–2987 (2006).
Cairns, J. Cancer and the immortal strand hypothesis. Genetics 174, 1069–1072 (2006).
Sotiropoulou, P. A., Candi, A. & Blanpain, C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells 26, 2964–2973 (2008).
Shinin, V., Gayraud-Morel, B., Gomes, D. & Tajbakhsh, S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat. Cell Biol. 8, 677–687 (2006).
Conboy, M. J., Karasov, A. O. & Rando, T. A. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 5, e102 (2007).
Karpowicz, P. et al. The germline stem cells of Drosophila melanogaster partition DNA non-randomly. Eur. J. Cell Biol. 88, 397–408 (2009).
Price, J., Turner, D. & Cepko, C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl Acad. Sci. USA 84, 156–160 (1987).
Axelrod, D. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys. J. 26, 557–573 (1979).
Dick, J. E. Retrovirus-mediated gene transfer into hematopoietic stem cells. Ann. N. Y. Acad. Sci. 507, 242–251 (1979).
Dzierzak, E. A, Papayannopoulou, T. & Mulligan, R. C. Lineage-specific expression of a human β-globin gene in murine bone marrow transplant recipients reconstituted with retrovirus-transduced stem cells. Nature 331, 35–41 (1988).
Golic, K. G. Site-specific recombination between homologous chromosomes in Drosophila. Science 252, 958–961 (1991).
Lakso, M. et al. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl Acad. Sci. USA 89, 6232–6236 (1992).
Harrison, D. A. & Perrimon, N. Simple and efficient generation of marked clones in Drosophila. Curr. Biol. 3, 424–433 (1993).
Margolis, J. & Spradling, A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development 121, 3797–3807 (1995).
Ohlstein, B. & Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006).
Soriano, P. Generalized lacZ expression with the Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Zhang, Y. V, Cheong, J., Ciapurin, N., McDermitt, D. J & Tumbar, T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell 5, 267–278 (2009).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Morris, R. J. et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 (2004).
Jaks, V. et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291–1299 (2008).
Barker, N. et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Greco, V. et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4, 155–169 (2009).
Jensen, P. et al. Redefining the serotonergic system by genetic lineage. Nat. Neurosci. 11, 417–419 (2008).
Kriegstein, A. & Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 (2008).
Klein, A. M., Nakagawa, T., Ichikawa, R., Yoshida, S. & Simons, B. D. Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell 7, 214–224 (2010).
Sulston, J. E., Schierenberg, E., White, J. G. & Thomson, J. N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983).
Alexandre, P., Reugels, A. M., Barker, D., Blanc, E. & Clarke, J. D. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat. Neurosci. 13, 673–679 (2010).
Celso, C. L. et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457, 92–96 (2009).
Xie, Y. et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature 457, 97–101 (2009).
Nakagawa, T., Sharma, M., Nabeshima, Y., Braun, R. E. & Yoshida, S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62–67 (2010).
Acknowledgements
We would like to thank our colleagues in the stem cell field whose ingenuity and creativity have developed these technologies for stem cell biology. In particular, we thank T. Tumbar (Cornell University, USA), H. Hock (Harvard Medical School, Harvard Stem Cell Institute and Cancer Center and Centre for Regenerative Medicine, Harvard University, USA), H. Clevers (Hubrecht Institute, the Netherlands), H. Snippert (Clevers Lab, Hubrecht Institute, the Netherlands), N. Barker (Hubrecht Institute, the Netherlands), Y-C. Hsu (Fuchs lab, Rockefeller University, USA) and J. Nowak (Rockefeller University, USA) for providing images. V.H. is a Pew Scholar in Biomedical Research and is funded by funded by the NIH (4R00AR054775) and the Connecticut Dept. Public Health (09SCAYALE30). E.F. is an HHMI investigator and receives support for her research on the identification and tracking of stem cells from the NIH (R01-AR050452) and New York State.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Fuchs, E., Horsley, V. Ferreting out stem cells from their niches. Nat Cell Biol 13, 513–518 (2011). https://doi.org/10.1038/ncb0511-513
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb0511-513
This article is cited by
-
CD9, a potential leukemia stem cell marker, regulates drug resistance and leukemia development in acute myeloid leukemia
Stem Cell Research & Therapy (2021)
-
IL-11 is essential in promoting osteolysis in breast cancer bone metastasis via RANKL-independent activation of osteoclastogenesis
Cell Death & Disease (2019)
-
Mesenchymal Stem Cells in the Musculoskeletal System: From Animal Models to Human Tissue Regeneration?
Stem Cell Reviews and Reports (2018)
-
Targeted knock-in of CreER T2 in zebrafish using CRISPR/Cas9
Cell and Tissue Research (2018)
-
Comparing adult renal stem cell identification, characterization and applications
Journal of Biomedical Science (2017)