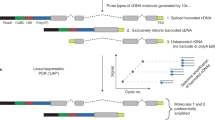
Abstract
Full-length RNA sequencing (RNA-Seq) has been applied to bulk tissue, cell lines and sorted cells to characterize transcriptomes1,2,3,4,5,6,7,8,9,10,11, but applying this technology to single cells has proven to be difficult, with less than ten single-cell transcriptomes having been analyzed thus far12,13. Although single splicing events have been described for ≤200 single cells with statistical confidence14,15, full-length mRNA analyses for hundreds of cells have not been reported. Single-cell short-read 3′ sequencing enables the identification of cellular subtypes16,17,18,19,20,21, but full-length mRNA isoforms for these cell types cannot be profiled. We developed a method that starts with bulk tissue and identifies single-cell types and their full-length RNA isoforms without fluorescence-activated cell sorting. Using single-cell isoform RNA-Seq (ScISOr-Seq), we identified RNA isoforms in neurons, astrocytes, microglia, and cell subtypes such as Purkinje and Granule cells, and cell-type-specific combination patterns of distant splice sites6,7,8,9,22,23. We used ScISOr-Seq to improve genome annotation in mouse Gencode version 10 by determining the cell-type-specific expression of 18,173 known and 16,872 novel isoforms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
BioProject
Sequence Read Archive
References
Sharon, D., Tilgner, H., Grubert, F. & Snyder, M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31, 1009–1014 (2013).
Au, K.F. et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proc. Natl. Acad. Sci. USA 110, E4821–E4830 (2013).
Oikonomopoulos, S., Wang, Y.C., Djambazian, H., Badescu, D. & Ragoussis, J. Benchmarking of the Oxford Nanopore MinION sequencing for quantitative and qualitative assessment of cDNA populations. Sci. Rep. 6, 31602 (2016).
Tilgner, H., Grubert, F., Sharon, D. & Snyder, M.P. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. USA 111, 9869–9874 (2014).
Koren, S. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30, 693–700 (2012).
Tilgner, H. et al. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat. Biotechnol. 33, 736–742 (2015).
Tilgner, H. et al. Microfluidic isoform sequencing shows widespread splicing coordination in the human transcriptome. Genome Res. 28, 231–242 (2018).
Bolisetty, M.T., Rajadinakaran, G. & Graveley, B.R. Determining exon connectivity in complex mRNAs by nanopore sequencing. Genome Biol. 16, 204 (2015).
Roy, C.K., Olson, S., Graveley, B.R., Zamore, P.D. & Moore, M.J. Assessing long-distance RNA sequence connectivity via RNA-templated DNA–DNA ligation. eLife 4, e03700 (2015).
Treutlein, B., Gokce, O., Quake, S.R. & Südhof, T.C. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc. Natl. Acad. Sci. USA 111, E1291–E1299 (2014).
Schreiner, D. et al. Targeted combinatorial alternative splicing generates brain region–specific repertoires of neurexins. Neuron 84, 386–398 (2014).
Karlsson, K. & Linnarsson, S. Single-cell mRNA isoform diversity in the mouse brain. BMC Genomics 18, 126 (2017).
Byrne, A. et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 8, 16027 (2017).
Song, Y. et al. Single-cell alternative splicing analysis with expedition reveals splicing dynamics during neuron differentiation. Mol. Cell 67, 148–161 (2017).
Shalek, A.K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Zeisel, A. et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Lake, B.B. et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016).
Zheng, G.X.Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 112, 7285–7290 (2015).
Jaitin, D.A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Pollen, A.A. et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 (2014).
Fededa, J.P. et al. A polar mechanism coordinates different regions of alternative splicing within a single gene. Mol. Cell 19, 393–404 (2005).
Fagnani, M. et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 8, R108 (2007).
Mecklenburg, N. et al. Growth and differentiation factor 10 (Gdf10) is involved in Bergmann glial cell development under Shh regulation. Glia 62, 1713–1723 (2014).
Koirala, S. & Corfas, G. Identification of novel glial genes by single-cell transcriptional profiling of Bergmann glial cells from mouse cerebellum. PLoS One 5, e9198 (2010).
Lein, E.S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Butts, T., Green, M.J. & Wingate, R.J.T. Development of the cerebellum: simple steps to make a 'little brain'. Development 141, 4031–4041 (2014).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Waterston, R.H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Tilgner, H. et al. Accurate identification and analysis of human mRNA isoforms using deep long read sequencing. G3 3, 387–397 (2013).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Mcmanus et al. Global analysis of trans-splicing in Drosophila. Proc. Natl. Acad. Sci. USA 107, 12975–12979 (2010).
Karolchik, D. et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770 (2014).
O'Leary, N.A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016).
Ge, K. et al. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc. Natl. Acad. Sci. USA 96, 9689–9694 (1999).
Fugier, C. et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 17, 720–725 (2011).
Karni, R. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 (2007).
Anvar, S.Y. et al. Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing. Genome Biol. 19, 46 (2018).
Vaquero-Garcia, J. et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife 5, e11752 (2016).
Li, Y.I. et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 50, 151–158 (2018).
Sharma, K. et al. Cell type– and brain region–resolved mouse brain proteome. Nat. Neurosci. 18, 1819–1831 (2015).
Kang, H.M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Satija, R., Farrell, J.A., Gennert, D., Schier, A.F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Wu, T.D. & Watanabe, C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Acknowledgements
This work used the Genomics Resources Core Facility and owes special thanks to J. Xiang and A. Wan. This work was supported by start-up funds (Weill Cornell Medicine) and a Leon Levy Fellowship in Neuroscience to H.U.T. as well as an R01 from the National Institute of Neurological Disorders and Stroke (1R01NS105477) to M.E.R.
Author information
Authors and Affiliations
Contributions
P.G.C., I.G., S.A.S. and H.U.T. devised the experiments. P.G.C., B.H., I.G., S.A.S., O.F. and W.L. performed the experiments. I.G., A.B.S. and H.U.T. devised the analyses. I.G., A.M., A.J., T.F., F.K. and H.U.T. performed the analyses. All of the authors discussed and interpreted the results throughout the project. I.G. and H.U.T. wrote the paper with inputs from all of the other authors. B.B., M.E.R. and H.U.T. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Figure S1
(A) Comparison of ScISOr-Seq approach to sorting methods. (B) Expression patterns of selected marker genes across each cell-type cluster.
Supplementary Figure 2 Figure S2: Illumina Short-read 3’seq statistics.
Distribution of (A) short reads (B) short-read UMIs and (C) genes per cell. (D) Number of total cells, clustered cells and unclassified cells. (E) Number of cells per cluster. Distribution of (F) short reads (G) short-read UMIs and (H) detected genes for each cell-type cluster.
Supplementary Figure 3 Figure S3: Comparison of single cell biological replicates.
Pairwise Jaccard distance between different identified cell-types calculated as the ratio of the number of genes in the intersection to the number of genes in the union: (A) Within replicate 1, (B) within replicate 2, C) replicate 1 vs. replicate 2. D) Fraction of cells each cell-type cluster contains in single cell replicates 1 and 2.
Supplementary Figure 4 Figure S4: Long-read statistics.
Distribution of (A) long reads (B) long-read UMIs and (C) genes per cell. (D) Number of cells >1,>10,>100,>250 long-reads (E) Dotplot and correlation between long-read UMIs and short-read UMIs per cell. Distribution of (F) long reads (G) long-read UMIs and (H) long-read detected genes for each cell-type cluster.
Supplementary Figure 5 Figure S5: PolyA and barcode identification statistics for ScISOr-Seq using Nanopore.
Detection of polyA tail is based on finding the first window of 30 bps with >=25 bps annotated as “A” or a “T” in the appropriate direction (A) Distribution of read length for reads with (red) and without (black) polyA tail for 1D2 pass (left), 1D2 failed (center) and 1D reads(right). (B) Histogram of polyA tail position for 1D2 pass (left), 1D2 failed(center) and 1D reads(right). We expect the polyA tail to begin at about 121 bps from the beginning of a read, however due to higher error rates in reads the detected position of polyA tail was fuzzy and therefore we found a wide spread of ~90bps around the expected position of ~121 bps unlike in PacBio reads. (C) Barplot for the percentage of reads with polyA tails (D) Barplot for the percentage of reads with a polyA tail for which a unique 10x cell barcode was found (E) Barplot for the percentage of reads with a polyA tail, for which multiple or 1-mismatch 10x cell barcode were found in 1D2 pass (green), 1D2 failed(purple) and 1D reads(gray).
Supplementary Figure 6 Figure S6: Mapping statistics.
(A) Distribution of read length for long-reads (B) Number of molecules submitted for mapping (left bar) and number (and percentage) of molecules that could be mapped to the mm10 genome using STARlong. (C) Number and percentage of molecules for which we could determine a single high-confidence-mapping (well-mapped, left bar) and those that did not overlap ribosomal RNA genes (right, percentage with respect to previous bar). (D) Number of molecules falling entirely into one intergenic, intronic or exonic region. Note that the definition of intergenic used here is based on the Gencode-vM10 annotation, which defines lncRNA genes, ribosomal RNA genes and many other kinds of short RNA genes as ”genes”. (E) Intron length distribution for introns in consensus-split-molecule-mappings, showing only introns of up to 1.5kb. The red dashed line (at 70bps) indicates a cutoff under which very few annotated human introns could be found (see reference 8), suggesting a minimal intron-size of this length, under which human introns might be difficult to process for the splicing machinery.
Supplementary Figure 7 Figure S7
(A) Distributions of coverage with astrocytic (obtained by immune-panning) short reads for junctions in the enhanced annotation that were exclusively observed in one cell type (indicated by name under the x-axis), with at least one observation in that cell type. (B) Distributions of coverage with astrocytic short reads for junctions in the enhanced annotation that were exclusively observed in one cell type (indicated by name under the x-axis), with at least three observations in that cell type. Horizontal red lines indicate the median value for junctions exclusively observed in astrocytic long-reads.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 2001 kb)
Supplementary Dataset 1
Complete Annotation of RNA Isoforms and their cell-type specific expression in the mammalian cerebellum from P1 mouse (ZIP 18439 kb)
Supplementary Dataset 2
Annotation of Novel RNA Isoforms, which are novel with respect to UCSC and RefSeq annotation, and their cell-type specific expression in the mammalian cerebellum from P1 mouse (ZIP 113 kb)
Supplementary Note 1
Supplementary note detailing methodology of defining trustworthy alignments to genes and detection of novel isoforms (PDF 199 kb)
Supplementary Code
R markdown detailing the single cell analysis pipeline (TXT 11 kb)
Rights and permissions
About this article
Cite this article
Gupta, I., Collier, P., Haase, B. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat Biotechnol 36, 1197–1202 (2018). https://doi.org/10.1038/nbt.4259
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4259
This article is cited by
-
Widespread changes in alternative splicing in developing and adult mouse brain
Nature Neuroscience (2024)
-
Neuron type-specific proteomics reveals distinct Shank3 proteoforms in iSPNs and dSPNs lead to striatal synaptopathy in Shank3B–/– mice
Molecular Psychiatry (2024)
-
Unique transcriptomes of sensory and non-sensory neurons: insights from Splicing Regulatory States
Molecular Systems Biology (2024)
-
Single-cell long-read sequencing-based mapping reveals specialized splicing patterns in developing and adult mouse and human brain
Nature Neuroscience (2024)
-
Near-gapless and haplotype-resolved apple genomes provide insights into the genetic basis of rootstock-induced dwarfing
Nature Genetics (2024)