Abstract
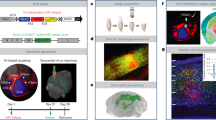
Neocortical areas communicate through extensive axonal projections, but the logic of information transfer remains poorly understood, because the projections of individual neurons have not been systematically characterized. It is not known whether individual neurons send projections only to single cortical areas or distribute signals across multiple targets. Here we determine the projection patterns of 591 individual neurons in the mouse primary visual cortex using whole-brain fluorescence-based axonal tracing and high-throughput DNA sequencing of genetically barcoded neurons (MAPseq). Projections were highly diverse and divergent, collectively targeting at least 18 cortical and subcortical areas. Most neurons targeted multiple cortical areas, often in non-random combinations, suggesting that sub-classes of intracortical projection neurons exist. Our results indicate that the dominant mode of intracortical information transfer is not based on ‘one neuron–one target area’ mapping. Instead, signals carried by individual cortical neurons are shared across subsets of target areas, and thus concurrently contribute to multiple functional pathways.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Harris, K. D. & Shepherd, G. M. G. The neocortical circuit: themes and variations. Nat. Neurosci. 18, 170–181 (2015)
Nakamura, H., Gattass, R., Desimone, R. & Ungerleider, L. G. The modular organization of projections from areas V1 and V2 to areas V4 and TEO in macaques. J. Neurosci. 13, 3681–3691 (1993)
Segraves, M. A. & Innocenti, G. M. Comparison of the distributions of ipsilaterally and contralaterally projecting corticocortical neurons in cat visual cortex using two fluorescent tracers. J. Neurosci. 5, 2107–2118 (1985)
Rockland, K. S. Collateral branching of long-distance cortical projections in monkey. J. Comp. Neurol. 521, 4112–4123 (2013)
Sincich, L. C. & Horton, J. C. Independent projection streams from macaque striate cortex to the second visual area and middle temporal area. J. Neurosci. 23, 5684–5692 (2003)
Yamashita, T. et al. Membrane potential dynamics of neocortical projection neurons driving target-specific signals. Neuron 80, 1477–1490 (2013)
Glickfeld, L. L., Andermann, M. L., Bonin, V. & Reid, R. C. Cortico-cortical projections in mouse visual cortex are functionally target specific. Nat. Neurosci. 16, 219–226 (2013)
Sato, T. R. & Svoboda, K. The functional properties of barrel cortex neurons projecting to the primary motor cortex. J. Neurosci. 30, 4256–4260 (2010)
Chen, J. L., Carta, S., Soldado-Magraner, J., Schneider, B. L. & Helmchen, F. Behaviour-dependent recruitment of long-range projection neurons in somatosensory cortex. Nature 499, 336–340 (2013)
Yamashita, T. & Petersen, C. C. H. Target-specific membrane potential dynamics of neocortical projection neurons during goal-directed behavior. eLife 5, e15798 (2016)
Movshon, J. A. & Newsome, W. T. Visual response properties of striate cortical neurons projecting to area MT in macaque monkeys. J. Neurosci. 16, 7733–7741 (1996)
Nassi, J. J. & Callaway, E. M. Parallel processing strategies of the primate visual system. Nat. Rev. Neurosci. 10, 360–372 (2009)
Andermann, M. L., Kerlin, A. M., Roumis, D. K., Glickfeld, L. L. & Reid, R. C. Functional specialization of mouse higher visual cortical areas. Neuron 72, 1025–1039 (2011)
Marshel, J. H. H., Garrett, M. E. E., Nauhaus, I. & Callaway, E. M. M. Functional specialization of seven mouse visual cortical areas. Neuron 72, 1040–1054 (2011). 8
Massé, I. O ., Régnier, P & Boire, D. in Axons and Brain Architecture (ed. Rockland, K. S. ) Ch. 5, 93 –116 (Academic, 2016)
Bullier, J. & Kennedy, H. Axonal bifurcation in the visual system. Trends Neurosci. 10, 205–210 (1987)
Economo, M. N. et al. A platform for brain-wide imaging and reconstruction of individual neurons. eLife 5, e10566 (2016)
Vogt Weisenhorn, D. M., Illing, R. B. & Spatz, W. B. Morphology and connections of neurons in area 17 projecting to the extrastriate areas MT and 19DM and to the superior colliculus in the monkey Callithrix jacchus. J. Comp. Neurol. 362, 233–255 (1995)
Ding, S.-L., Van Hoesen, G. & Rockland, K. S. Inferior parietal lobule projections to the presubiculum and neighboring ventromedial temporal cortical areas. J. Comp. Neurol. 425, 510–530 (2000)
Zingg, B. et al. Neural networks of the mouse neocortex. Cell 156, 1096–1111 (2014)
Oh, S. W. et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014)
Wang, Q. & Burkhalter, A. Area map of mouse visual cortex. J. Comp. Neurol. 502, 339–357 (2007)
Ragan, T. et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258 (2012)
Osten, P. & Margrie, T. W. Mapping brain circuitry with a light microscope. Nat. Methods 10, 515–523 (2013)
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007)
D’Souza, R. D., Meier, A. M., Bista, P., Wang, Q. & Burkhalter, A. Recruitment of inhibition and excitation across mouse visual cortex depends on the hierarchy of interconnecting areas. eLife 5, e19332 (2016)
Yang, W., Carrasquillo, Y., Hooks, B. M., Nerbonne, J. M. & Burkhalter, A. Distinct balance of excitation and inhibition in an interareal feedforward and feedback circuit of mouse visual cortex. J. Neurosci. 33, 17373–17384 (2013)
Zhuang, J. et al. An extended retinotopic map of mouse cortex. eLife 6, e18372 (2017)
Gong, H. et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 7, 12142 (2016)
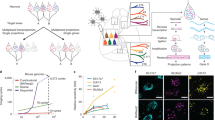
Kebschull, J. M. et al. High-throughput mapping of single-neuron projections by sequencing of barcoded RNA. Neuron 91, 975–987 (2016)
Wang, Q., Sporns, O. & Burkhalter, A. Network analysis of corticocortical connections reveals ventral and dorsal processing streams in mouse visual cortex. J. Neurosci. 32, 4386–4399 (2012)
Smith, I. T., Townsend, L. B., Huh, R., Zhu, H. & Smith, S. L. Stream-dependent development of higher visual cortical areas. Nat. Neurosci. 20, 200–208 (2017)
Murakami, T., Matsui, T. & Ohki, K. Functional segregation and development of mouse higher visual areas. J. Neurosci. 37, 9424–9437 (2017)
Pecka, M., Han, Y., Sader, E. & Mrsic-Flogel, T. D. Experience-dependent specialization of receptive field surround for selective coding of natural scenes. Neuron 84, 457–469 (2014)
Pologruto, T. A., Sabatini, B. L. & Svoboda, K. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13 (2003)
Mayerich, D., Abbott, L. & McCormick, B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J. Microsc. 231, 134–143 (2008)
Niedworok, C. J. et al. aMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data. Nat. Commun. 7, 11879 (2016)
Klein, S., Staring, M., Murphy, K., Viergever, M. A. & Pluim, J. P. elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205 (2010)
Kim, Y. et al. Whole-brain mapping of neuronal activity in the learned helplessness model of depression. Front. Neural Circuits 10, 3 (2016)
Renier, N. et al. Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016)
Roth, M. M. et al. Thalamic nuclei convey diverse contextual information to layer 1 of visual cortex. Nat. Neurosci. 19, 299–307 (2016)
Bryan, W. P. & Byrne, R. H. A calcium chloride solution, dry-ice, low temperature bath. J. Chem. Educ. 47, 361 (1970)
Morris, J., Singh, J. M. & Eberwine, J. H. Transcriptome analysis of single cells. J. Vis. Exp. 50, e2634 (2011)
Acknowledgements
We thank A. Juavinett, L. Huang, S. Hofer and P. Znamenskiy for comments on the manuscript. This study was funded by National Institutes of Health (5RO1NS073129 and 5RO1DA036913 to A.M.Z.); Brain Research Foundation (BRF-SIA-2014-03 to A.M.Z.); IARPA (MICrONS D16PC0008 to A.M.Z.); Simons Foundation (382793/SIMONS to A.M.Z.); Paul Allen Distinguished Investigator Award (to A.M.Z.); PhD fellowship from the Boehringer Ingelheim Fonds (to J.M.K.); PhD fellowship from the Genentech Foundation (to J.M.K.); National Natural Science Foundation of China (NSFC 31600847 to Y.H.); European Research Council (NeuroV1sion 616509 to T.D.M.-F.), and Swiss National Science Foundation (SNSF 31003A_169802 to T.D.M.-F.).
Author information
Authors and Affiliations
Contributions
Y.H. generated the dataset for fluorescence-based axonal tracing. D.C. and Y.H. traced the cells. R.A.A.C. analysed the serial two-photon imaging data and axonal projection patterns. J.M.K. and F.I. collected the MAPseq dataset. J.M.K. and A.M.Z. performed the analysis of projection patterns. J.M.K., T.D.M.-F. and A.M.Z. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks M. Helmstaedter, O. Sporns and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Single-neuron tracing protocol efficiently fills axons projecting to the ipsilateral striatum.
We retrogradely labelled striatum-projecting cells by stereotactically injecting cholera toxin subunit B conjugated to AlexaFluor 594 or PRV-Cre into the visual striatum of wild-type mice or tdTomato-reporter mice (Ai14, Jax), respectively (magenta). With visual guidance of two-photon microscopy, we electroporated single retrogradely labelled cells in V1 with a GFP-expressing plasmid (cyan). a, Coronal, maximum intensity projections of visual striatum. Scale bar, 1 mm. b, Higher magnification image of the visual striatum. Scale bar, 0.2 mm. c, Single-channel images of the same axonal arborization as in b. d, Coronal maximum intensity projection containing V1. Scale bar, 1 mm. e, Higher magnification image of V1. Scale bar, 0.2 mm. f, Single-channel images of V1. Scale bars, 0.2 mm. g, Horizontal projections in the Allen Reference Atlas space of eight retrogradely labelled and electroporated cells. Cell ID numbers are indicated at the top right of each image. Scale bars, 1 mm. Note that one additional cell was retrogradely labelled and electroporated, which revealed its axonal projection to the striatum, but it is not shown because the brain was too distorted to allow accurate registration to the Allen Reference Atlas.
Extended Data Figure 2 Some axonal branches terminate abruptly without arborizing, whereas other branches of the same neuron arborize extensively within different target areas and appear to be completely filled.
a, Horizontal view of a representative cell in the Allen Reference Atlas space. The abrupt termination is labelled with a purple square. n = 28 abruptly terminating cells. b, The abrupt termination of the example cell shown as a maximum z projection (left) and in the individual z sections (right). c, Two normal terminations of the same cell, shown as a maximum z projection (left) and in two colour-coded series of z sections (right). d, Distance of abrupt termination from cell body versus distance of furthest regular termination of the same cell. Dashed line indicates the unity line. e, The distribution of target numbers of all projection neurons without abrupt terminations (as shown in the main figures; left), of projection cells with abrupt terminations (middle) and of all projection neurons (no abrupt terminations and abrupt terminations; right). f, To test the effect of false negatives on our analyses, we simulated the random loss or gain of projections from the MAPseq dataset, while maintaining overall area projection probabilities. n = 553 neurons; 400 repeats. P values based on a binomial test for all six projection motifs determined as significantly over- or underrepresented in our dataset are plotted after removing (dropfraction <1) or adding (dropfraction >1) connections. Mean (black line) and s.d. (shaded area) are indicated.
Extended Data Figure 3 Images of traced layer-2/3 V1 neurons.
Horizontal views of the Allen Reference Atlas space are shown, and cell ID numbers are indicated at the top right of each image. Scale bars, 1 mm.
Extended Data Figure 4 Images of traced layer-2/3 V1 neurons, continued.
Horizontal views of the Allen Reference Atlas space are shown, and cell ID numbers are indicated at the top right of each image. Scale bars, 1 mm.
Extended Data Figure 5 Individual neurons in higher visual areas project to more than one target area.
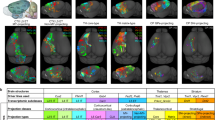
a, All traced neurons with cell bodies not in V1. Brain area identity is colour coded as in Fig. 1. Cell identity is indicated at the top right of each image. Scale bars, 1 mm. b, Histogram of the number of target areas per cell.
Extended Data Figure 6 Density of axonal innervation by area and layer of V1 layer-2/3 projection neurons.
a, Total axon length plotted as a function of the number of targets innervated by every V1 projection neuron. b, Axon length in area LM, PM or POR plotted as a function of the total number of targets innervated by each neuron projection to the respective area. c–h, The axons of V1 neurons in target areas most densely innervate layers 2/3 and 5, with some density in layer 1, but less in layers 4 and 6, often recapitulating the laminar axonal profile within V1. Coronal views of each area are shown in Allen Reference Atlas space (left) and axonal arborizations of each neuron innervating the area are colour coded. Scale bars, 200 μm. A histogram of the laminar innervation is shown (right). Note that cells with abrupt terminations outside the shown area were included in this analysis. Areas depicted are V1 (c), AL (d), LI (e), LM (f), PM (g) and POR (h). White-matter axons are not shown.
Extended Data Figure 7 Conclusions from fluorescence-based single-neuron tracing data hold true when the analysis is restricted to subset of target areas.
a, The projection patterns of reconstructed GFP-filled neurons when only the six target areas (LI, LM, AL, PM, AM and RL) are considered. Projection strengths are normalized to the maximum projection of each neuron, and only neurons projecting to at least one target area are shown. b, The distribution of target area numbers per projection neuron. c, The fraction of all cells projecting to each target area. d, The fraction of dedicated input per area. e, The number of times each binarized projection motif is observed. f, The fraction of broadcasting cells as a function of the minimum projection strength (relative to the primary target) that each area needs to receive to be considered a target. g, The fraction of broadcasting cells as a function of increasing buffer zones between areas within which axons are ignored, assuming a minimum projection of 1 mm of axon per target area. h, The fraction of broadcasting cells as a function of the minimal amount of axon per area for it to be considered a target, assuming buffer zones of 100-μm width. Black arrowheads indicate chosen buffer zones and minimal projection for analysis in the paper.
Extended Data Figure 8 Location of cell bodies in V1 as a function of their projection targets.
a–l, Horizontal views of Allen Reference Atlas space are shown. The location of all traced V1 neurons are indicated as circles (cells with no abrupt terminations) or squares (cells with abrupt terminations). In each plot, the cells projecting to the highlighted higher visual area are coloured in solid blue. Target areas considered are A (a), AL (b), AM (c), ECT (d), LI (e), LM (f), P (g), PER (h), PM (i), POR (j), RL (k) and TEA (l). m, n, Quantification of cell body location in the rostrocaudal (m) and mediolateral (n) direction. Dotted lines indicate expected number of cells based on a bootstrapping procedure, for which we randomly selected neurons from the available positions to project to each area and repeated the process 10,000 times. P values were derived from the bootstrapping probability distribution and are indicated for projection targets significantly deviating from this expectation (α = 0.05). P values below 10−4 are not exact and are therefore indicated as a range.
Extended Data Figure 9 MAPseq dissection strategy.
We identified the to-be-dissected higher visual areas by performing intrinsic imaging of visual cortex in response to stimuli at different positions in the contralateral visual field and mapping the resulting changes in intrinsic signals. a, A representative retinotopic map, with responses to the two 25° visual stimuli pseudocoloured in green and magenta (stimulus 1 position: 90° azimuth, 20° elevation; stimulus 2 position: 60° azimuth, 20° elevation). On the basis of this map, we fluorescently labelled retinotopically matched positions in the to-be-dissected cortical areas with a DiI stab (white circles). Putative borders between the higher visual areas are indicated with dashed lines for orientation. Scale bar, 1 mm. n = 4 mice. b, The MAPseq virus injection site is discernible in consecutive frozen 180-μm thick coronal sections, using GFP fluorescence. Scale bars, 1 mm. c, DiI injections targeted to matched retinotopic positions in six target areas identified by intrinsic signal imaging. DiI epifluorescence images of each 180-μm thick slice are shown, and dissected areas are labelled. Scale bars, 1 mm.
Extended Data Figure 10 Clustering of MAPseq data and data summary.
a, b, Gap (a) and silhouette criteria (b) for k-means clustering of the MAPseq neurons as a function of the number of clusters. Black arrow heads indicate chosen number of clusters (k = 8). c, d, Centroids for alternative, near-optimal cluster number choices with k = 3 (c) and k = 5 (d). e, Hierarchical clustering results of the MAPseq dataset using a cosine distance metric. c–e, Colour intensity indicates projection strengths. f, g, Summary of single-neuron projections from V1. f, Cells targeting single higher visual areas (dedicated projection neurons) comprise the minority of layer-2/3 V1 projection neurons. Among the areas analysed by MAPseq, dedicated projection neurons predominantly innervate cortical areas LM or PM. g, Cells projecting to two or more areas (broadcasting projection neurons) are the dominant mode of information transfer from V1 to higher visual areas. In the six areas analysed by MAPseq, broadcasting neurons innervate combinations of target areas in a non-random manner, including those that are more or less abundant than expected by chance. Line width indicates the absolute abundance of each projection type as observed in the MAPseq dataset.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-4, Supplementary Table 1 and Supplementary References. (PDF 395 kb)
Rights and permissions
About this article
Cite this article
Han, Y., Kebschull, J., Campbell, R. et al. The logic of single-cell projections from visual cortex. Nature 556, 51–56 (2018). https://doi.org/10.1038/nature26159
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature26159
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.