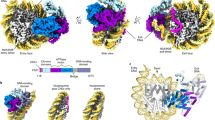
Abstract
Chromatin-remodelling factors change nucleosome positioning and facilitate DNA transcription, replication, and repair1. The conserved remodelling factor chromodomain-helicase-DNA binding protein 1(Chd1)2 can shift nucleosomes and induce regular nucleosome spacing3,4,5. Chd1 is required for the passage of RNA polymerase IIthrough nucleosomes6 and for cellular pluripotency7. Chd1 contains the DNA-binding domains SANT and SLIDE, a bilobal motor domain that hydrolyses ATP, and a regulatory double chromodomain. Here we report the cryo-electron microscopy structure of Chd1 from the yeast Saccharomyces cerevisiae bound to a nucleosome at a resolution of 4.8 Å. Chd1 detaches two turns of DNA from the histone octamer and binds between the two DNA gyres in a state poised for catalysis. The SANT and SLIDE domains contact detached DNA around superhelical location (SHL) −7 of the first DNA gyre. The ATPase motor binds the second DNA gyre at SHL +2 and is anchored to the N-terminal tail of histone H4, as seen in a recent nucleosome–Snf2 ATPase structure8. Comparisons with published results9 reveal that the double chromodomain swings towards nucleosomal DNA at SHL +1, resulting in ATPase closure. The ATPase can then promote translocation of DNA towards the nucleosome dyad, thereby loosening the first DNA gyre and remodelling the nucleosome. Translocation may involve ratcheting of the two lobes of the ATPase, which is trapped in a pre- or post-translocation state in the absence8 or presence, respectively, of transition state-mimicking compounds.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Narlikar, G. J., Sundaramoorthy, R. & Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503 (2013)
Delmas, V., Stokes, D. G. & Perry, R. P. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc. Natl Acad. Sci. USA 90, 2414–2418 (1993)
Lieleg, C. et al. Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density. Mol. Cell. Biol. 35, 1588–1605 (2015)
Hughes, A. L. & Rando, O. J. Comparative genomics reveals Chd1 as a determinant of nucleosome spacing in vivo. G3 (Bethesda) 5, 1889–1897 (2015)
Lusser, A., Urwin, D. L. & Kadonaga, J. T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12, 160–166 (2005)
Skene, P. J., Hernandez, A. E., Groudine, M. & Henikoff, S. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife 3, e02042 (2014)
Gaspar-Maia, A. et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868 (2009)
Liu, X., Li, M., Xia, X., Li, X. & Chen, Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445 (2017)
Nodelman, I. M. et al. Interdomain communication of the Chd1 chromatin remodeler across the DNA gyres of the nucleosome. Mol. Cell 65, 447–459.e6 (2017)
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998)
Vasudevan, D., Chua, E. Y. D. & Davey, C. A. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010)
Hauk, G., McKnight, J. N., Nodelman, I. M. & Bowman, G. D. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol. Cell 39, 711–723 (2010)
Sharma, A., Jenkins, K. R., Héroux, A. & Bowman, G. D. Crystal structure of the chromodomain helicase DNA-binding protein 1 (Chd1) DNA-binding domain in complex with DNA. J. Biol. Chem. 286, 42099–42104 (2011)
Mohanty, B., Helder, S., Silva, A. P. G., Mackay, J. P. & Ryan, D. P. The chromatin remodelling protein CHD1 contains a previously unrecognised C-terminal helical domain. J. Mol. Biol. 428, 4298–4314 (2016)
Sundaramoorthy, R. et al. Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes. eLife 6, e22510–e22528 (2017)
Sinha, K. K., Gross, J. D. & Narlikar, G. J. Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Science 355, eaaa3761 (2017)
McKnight, J. N., Jenkins, K. R., Nodelman, I. M., Escobar, T. & Bowman, G. D. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol. Cell. Biol. 31, 4746–4759 (2011)
Bednar, J. et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell 66, 384–397.e8 (2017)
Gu, M. & Rice, C. M. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl Acad. Sci. USA 107, 521–528 (2010)
Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S. & Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125, 287–300 (2006)
Huang, S. et al. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 31, 4164–4170 (2012)
Singleton, M. R., Dillingham, M. S. & Wigley, D. B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 (2007)
Dürr, H., Körner, C., Müller, M., Hickmann, V. & Hopfner, K.-P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121, 363–373 (2005)
Saha, A., Wittmeyer, J. & Cairns, B. R. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 12, 747–755 (2005)
Saikrishnan, K., Powell, B., Cook, N. J., Webb, M. R. & Wigley, D. B. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell 137, 849–859 (2009)
Hopfner, K.-P. & Michaelis, J. Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr. Opin. Struct. Biol. 17, 87–95 (2007)
Wigley, D. B. & Bowman, G. D. A glimpse into chromatin remodeling. Nat. Struct. Mol. Biol. 24, 498–500 (2017)
Le Gallo, M. et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 44, 1310–1315 (2012)
Clapier, C. R., Längst, G., Corona, D. F., Becker, P. B. & Nightingale, K. P. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21, 875–883 (2001)
Nodelman, I. M. et al. The Chd1 chromatin remodeler can sense both entry and exit sides of the nucleosome. Nucleic Acids Res. 44, 7580–7591 (2016)
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009)
Leonard, J. D. & Narlikar, G. J. A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler. Mol. Cell 57, 850–859 (2015)
Yan, L., Wang, L., Tian, Y., Xia, X. & Chen, Z. Structure and regulation of the chromatin remodeller ISWI. Nature 540, 466–469 (2016)
Xu, Y. et al. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat. Commun. 8, 15741 (2017)
Vos, S. M. et al. Architecture and RNA binding of the human negative elongation factor. eLife 5, e14981 (2016)
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999)
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2003)
Maskell, D. P. et al. Structural basis for retroviral integration into nucleosomes. Nature 523, 366–369 (2015)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Scheres, S. H. W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012)
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016)
Plaschka, C. et al. Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996)
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Thomsen, N. D. & Berger, J. M. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139, 523–534 (2009)
Schrödinger, LLC. The PyMOL Molecular Graphics System, v.1.8 (Schrödinger, 2015)
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013)
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009)
Acknowledgements
We thank past and present members of the Cramer laboratory including F. Fischer, R. Kohler, S. Neyer, D. Tegunov, and Y. Xu. We thank the members of the Halic laboratory for Xenopus laevis histone expression plasmids, a plasmid containing the Widom 601 sequence and initial advice on histone purification. S.M.V. was supported by an EMBO Long-Term-Fellowship (ALTF 745-2014). P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP1935), the European Research Council Advanced Investigator Grant TRANSREGULON (grant agreement No. 693023), and the Volkswagen Foundation.
Author information
Authors and Affiliations
Contributions
L.F. designed and carried out experiments and performed cryo-EM data acquisition and analysis. S.M.V. developed the protein expression strategy, performed baculovirus production, and insect cell expression. C.W. assisted with cryo-EM grid preparation and data collection. P.C. designed and supervised research. L.F. and P.C. interpreted the data and wrote the manuscript, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
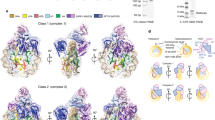
Extended Data Figure 1 Cryo-EM structure determination and analysis.
a, Formation of the nucleosome–Chd1–FACT–Paf1C complex. SDS–PAGE of peak fraction used for cryo-EM grid preparation containing Chd1, FACT subunits, Paf1C subunits, and histones. The identity of the bands was confirmed by mass spectrometry. For gel source data, see Supplementary Fig. 1. b, Representative cryo-EM micrograph of data collection. c, 2D class averages contain nucleosome-like shapes. d, Sorting and classification tree used to reconstruct the nucleosome–Chd1 particle at 4.8 Å resolution. Steps 1 and 2 of batch 1 global classification are shown representatively for all three batches.
Extended Data Figure 2 Quality of the nucleosome–Chd1 structure.
a, Overall fit of the nucleosome–Chd1 structure to the electron density. Two views are depicted as in Fig. 1b, c. b–f, Electron density (grey mesh) for various Chd1 domains reveals secondary structure and a good fit for DNA (SHL −4 to SHL +7). g, Superposition of the histone octamer core with canonical octamer core (PDB code 3LZ0). The canonical octamer core is rendered in grey. h, Nucleosome–Chd1 reconstruction coloured according to local resolution43. i, Angular distribution of particles. Red dots indicate the presence of at least one particle image assigned within ±1°. Shading from white to black indicates the density of particle images at a given orientation. j, Estimation of the average resolution. The dark blue line indicates the Fourier shell correlation (FSC) between the half maps of the reconstruction. The dotted light blue line indicates the Fourier shell correlation between the derived model and the reconstruction. Resolutions are given for the FSC 0.143 and the FSC 0.5 criteria. The dotted lines show the Fourier shell correlation between the derived Chd1 domains and the corresponding masked regions.
Extended Data Figure 3 Chd1–DNA interactions and Chd1 interaction interfaces.
a, Overview of Chd1–DNA interactions. b, Contact of chromo-wedge with DNA at SHL +1. c, Secondary DNA contacts of ATPase. Contact of motif Ib with first DNA gyre around SHL −6. d, Modelling linear B-DNA (orange) onto the ATPase motor in the nucleosome–Chd1 structure leads to a clash with the double chromodomain (purple). B-DNA was superimposed onto nucleosomal DNA at SHL +2. e, ADP·BeF3 binds in the active site of the Chd1 ATPase motor. Electron density is shown for ADP·BeF3, motif I (Walker A, P-loop, residues 403–410), motif II (Walker B, residues 510–515), and the arginine fingers (R804 and R807). Motifs I and II are shown in ribbon representation. ADP·BeF3 and the arginine finger residues are shown as sticks. The density for ADP is strong, whereas the density for BeF3− is weaker and thus we cannot formally rule out that BeF3− is not bound or shows only partial occupancy. f, Contact of W793 with the phosphate backbone of the guide strand at SHL +2. Electron density is shown as a grey mesh. Side chain of W793 is shown as a stick representation. g, Interface between the double chromodomain and the SANT and SLIDE domains of the DNA binding region. Chd1 domains are coloured as in Fig. 1a. h, Sequence of the Widom 601 sequence with 63 bp of extranucleosomal DNA.
Extended Data Figure 4 ATPase conservation and histone H4 tail binding.
a, Chd1 binds the N-terminal tail of histone H4 (green) with ATPase lobe 2 (surface representation coloured according to electrostatic surface potential; red, negative; white, neutral; blue, positive). The view is the inverse of that in Fig. 1b (that is, after a 180° rotation). b, Chd1 ATPase activity results in DNA translocation towards the octamer dyad, loosening DNA gyre 1 and triggering nucleosome remodelling. c, Sequence alignment of ATPase regions in S. cerevisiae (Sc) Chd1 (356–883), ScIsw1 (177–689), ScSnf2 (746–1270), Homo sapiens (Hs) Chd4 (703–1233), Drosophila melanogaster (Dm) Mi-2 (707–1231), and Sulfolobus solfataricus (Sso) Rad54 (423–802). Arginine ‘fingers’ of ScChd1 (R804 and R807) are indicated and ATPase motifs are underlined. Sequence coloured according to identity. Darker shades of blue indicate higher conservation, whereas lighter shades of blue indicate less conservation. Alignment was generated with MAFFT51 and visualized using JalView52.
Supplementary information
Supplementary Figure 1
This file contains the uncropped scan with size marker indication. The region presented in Extended Data Figure 1a is indicated by rectangle with dashed lines. (PDF 236 kb)
Overview of the nucleosome-Chd1 structure
A 3D overview of the nucleosome-Chd1 structure fitted into the cryo-EM electron density. (MOV 13340 kb)
Structural changes and ATPase activation
The video first shows DNA detachment, then swinging of the double chromodomain onto nucleosomal DNA, and then ATPase closure. (MP4 15262 kb)
Model for DNA translocation by the Chd1 ATPase motor
The video shows a model for DNA translocation by the Chd1 ATPase motor on B-DNA. For details see text. (MP4 7238 kb)
Rights and permissions
About this article
Cite this article
Farnung, L., Vos, S., Wigge, C. et al. Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017). https://doi.org/10.1038/nature24046
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24046
This article is cited by
-
Energy-driven genome regulation by ATP-dependent chromatin remodellers
Nature Reviews Molecular Cell Biology (2024)
-
Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking
Nature Communications (2024)
-
Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1
Nature Communications (2024)
-
Driving forces behind phase separation of the carboxy-terminal domain of RNA polymerase II
Nature Communications (2023)
-
Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.