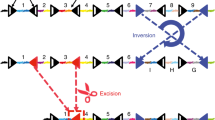
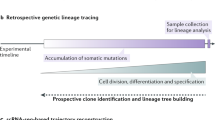
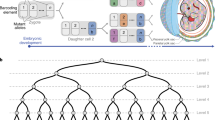
Abstract
Developmental deconvolution of complex organs and tissues at the level of individual cells remains challenging. Non-invasive genetic fate mapping1 has been widely used, but the low number of distinct fluorescent marker proteins limits its resolution. Much higher numbers of cell markers have been generated using viral integration sites2, viral barcodes3, and strategies based on transposons4 and CRISPR–Cas9 genome editing5; however, temporal and tissue-specific induction of barcodes in situ has not been achieved. Here we report the development of an artificial DNA recombination locus (termed Polylox) that enables broadly applicable endogenous barcoding based on the Cre–loxP recombination system6,7. Polylox recombination in situ reaches a practical diversity of several hundred thousand barcodes, allowing tagging of single cells. We have used this experimental system, combined with fate mapping, to assess haematopoietic stem cell (HSC) fates in vivo. Classical models of haematopoietic lineage specification assume a tree with few major branches. More recently, driven in part by the development of more efficient single-cell assays and improved transplantation efficiencies, different models have been proposed, in which unilineage priming may occur in mice and humans at the level of HSCs8,9,10. We have introduced barcodes into HSC progenitors in embryonic mice, and found that the adult HSC compartment is a mosaic of embryo-derived HSC clones, some of which are unexpectedly large. Most HSC clones gave rise to multilineage or oligolineage fates, arguing against unilineage priming, and suggesting coherent usage of the potential of cells in a clone. The spreading of barcodes, both after induction in embryos and in adult mice, revealed a basic split between common myeloid–erythroid development and common lymphocyte development, supporting the long-held but contested view of a tree-like haematopoietic structure.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kretzschmar, K. & Watt, F. M. Lineage tracing. Cell 148, 33–45 (2012)
Keller, G., Paige, C., Gilboa, E. & Wagner, E. F. Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature 318, 149–154 (1985)
Gerrits, A. et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood 115, 2610–2618 (2010)
Sun, J. et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014)
McKenna, A . et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016)
Sternberg, N. & Hamilton, D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150, 467–486 (1981)
Rajewsky, K. et al. Conditional gene targeting. J. Clin. Invest. 98, 600–603 (1996)
Yamamoto, R. et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 154, 1112–1126 (2013)
Notta, F . et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116 (2016)
Velten, L. et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281 (2017)
Hoess, R., Wierzbicki, A. & Abremski, K. Formation of small circular DNA molecules via an in vitro site-specific recombination system. Gene 40, 325–329 (1985)
Junker, J. P. et al. Massively parallel clonal analysis using CRISPR/Cas9 induced genetic scars. Preprint available at http://biorxiv.org/content/early/2017/01/04/056499 (2017)
Rybtsov, S., Ivanovs, A., Zhao, S. & Medvinsky, A. Concealed expansion of immature precursors underpins acute burst of adult HSC activity in foetal liver. Development 143, 1284–1289 (2016)
Busch, K. et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015)
Akashi, K., Traver, D., Miyamoto, T. & Weissman, I. L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000)
Terszowski, G. et al. Prospective isolation and global gene expression analysis of the erythrocyte colony-forming unit (CFU-E). Blood 105, 1937–1945 (2005)
Paul, F. et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677 (2015)
Perié, L., Duffy, K. R., Kok, L., de Boer, R. J. & Schumacher, T. N. The branching point in erythro-myeloid differentiation. Cell 163, 1655–1662 (2015)
Sawai, C. M. et al. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity 45, 597–609 (2016)
Schoedel, K. B. et al. The bulk of the hematopoietic stem cell population is dispensable for murine steady-state and stress hematopoiesis. Blood 128, 2285–2296 (2016)
Sheikh, B. N. et al. MOZ (KAT6A) is essential for the maintenance of classically defined adult hematopoietic stem cells. Blood 128, 2307–2318 (2016)
Weber, T. S. et al. Site-specific recombinatorics: in situ cellular barcoding with the Cre Lox system. BMC Syst. Biol. 10, 43 (2016)
Weissman, T. A. & Pan, Y. A. Brainbow: new resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics 199, 293–306 (2015)
Dick, J. E., Magli, M. C., Huszar, D., Phillips, R. A. & Bernstein, A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell 42, 71–79 (1985)
Kawamoto, H., Ikawa, T., Masuda, K., Wada, H. & Katsura, Y. A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol. Rev. 238, 23–36 (2010)
Ventura, A. et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007)
Pei, W. et al. Protocol for the use of Polylox—endogenous barcoding for high resolution in vivo lineage tracing. Protoc. Exch.http://dx.doi.org10.1038/protex.2017.092 (2017)
Pettitt, S. J. et al. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods 6, 493–495 (2009)
Hooper, M., Hardy, K., Handyside, A., Hunter, S. & Monk, M. HPRT-deficient (Lesch–Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326, 292–295 (1987)
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999)
Luche, H., Weber, O., Nageswara Rao, T., Blum, C. & Fehling, H. J. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37, 43–53 (2007)
Zhang, Y. et al. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 24, 543–548 (1996)
Chen, K. et al. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc. Natl Acad. Sci. USA 106, 17413–17418 (2009)
Acknowledgements
We thank J. Muehling, S. Oh, C. Heiner, P. Lobb, R. Lleras and C. Koenig (Pacific Biosciences), E. Hobeika (Ulm), S. Schäfer, T. Grünzinger (DKFZ), K. Reifenberg, M. Socher, A. Frenznick (Animal Facility DKFZ), F. v. der Hoeven, U. Kloz (Transgenic Service DKFZ), and N. Diessl, C. Previti and S. Wiemann (Genomics & Proteomics Core Facilities DKFZ) for help, and R. Hoess, H. Glimm and K. Rajewsky for discussions. T.H. is supported by CellNetworks, DKFZ core funding and e:Bio BMBF project FKZ 0316182B (SB-Epo); T.B.F. and H.-R.R. are supported by Transregio 156-A07; H.-R.R. is supported by SFB 873-B11, ERC Advanced Grant 742883, and DKFZ core funding.
Author information
Authors and Affiliations
Contributions
W.P., T.B.F., D.P., K.B., K.K. and N.D. performed experiments; W.P. and T.B.F. generated Polylox; X.W. and D.P. decoded Polylox; J.R. and T.H. performed barcode calculations and mathematical modelling; K.B. generated mice; I.R. suggested in vitro barcode testing; C.Q. sequenced barcodes, supported by W.C. and S.S.; S.W. provided sequencing considerations; T.H. and H.-R.R. supervised the study and wrote the paper, with input from T.B.F., and H.-R.R. conceived the study.
Corresponding authors
Ethics declarations
Competing interests
The German Cancer Research Center has filed an international patent application entitled ‘Genetic random DNA barcode generator for in vivo cell tracing’ (PCT/EP2016/065932). H.-R.R., T.B.F. and W.P. are listed as inventors.
Additional information
Reviewer Information Nature thanks S. Morrison, S. Orkin and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Generation of the Rosa26Polylox locus and experimental procedures for barcode detection and analysis.
a, Gene targeting of Polylox DNA into the Rosa26 locus in ES cells; shown are the wild type Rosa26 locus (top), targeting vector (middle) and targeted Rosa26Polylox locus (bottom). Southern blot (insert) (Supplementary Fig. 1b) from genomic tail DNA of control Rosa26+/+ and Rosa26RFP/+ (ref. 31) mice, and from three Rosa26Polylox/+ ES cell clones shows restriction fragments corresponding to wild-type (5.8 kb) or targeted (4.8 kb) loci. b, Kinetics of Polylox locus recombination after treatment of Rosa26Polylox MerCreMer ES cells with 4-hydroxy-tamoxifen (4-OHT) at 0 h (Supplementary Fig. 1c). c, Rosa26Polylox MerCreMer ES cells were left untreated and followed for 27 days (left), or pulsed with 4-OHT for 3 h and chased over 34 days (right) (Supplementary Fig. 1d). d, Workflow from cell isolation to Polylox barcode detection. Cell populations of interest were isolated by cell sorting, genomic DNA was purified and the Polylox cassette was amplified by PCR, and the fragments (see Methods) were sequenced by SMRT sequencing using Pacific Biosciences instruments. Raw data were processed with the accompanying software package to obtain the circular consensus sequences (CCS). Subsequently, CCS reads were filtered for reads containing complete Polylox sequences. Next, we aligned the barcode segments to the CCS reads and determined the order and orientation of the segments to retrieve the recombined Polylox barcodes (see Methods). Finally, CCS reads with incomplete segment alignment (X), or illegitimate segment orders (for example, segment duplications) were filtered out and removed from further analysis.
Extended Data Figure 2 Example of a complex CCS DNA sequence and its corresponding Polylox barcode.
a, Schematic drawing of the unrecombined Polylox cassette, and an experimentally found recombined barcode. b, Full nucleotide sequence of one CCS read. In 5′ to 3′ orientation, the DNA sequence is organized into intervening loxP sites and the annotated barcode blocks (‘barcode alphabet’). Numbers and letters refer to the segments shown in a. c, Proportions of unrecombined (blue) and recombined (red) sequence reads in granulocytes (Gr), B cells (B2), CD4 T cells (CD4), and CD8 T cells (CD8) from adult Rosa26PolyloxTie2MCM mice without tamoxifen (TAM) treatment (top) and adult Rosa26Polylox (middle) and adult Rosa26PolyloxTie2MCM (bottom) mice, each treated with tamoxifen as embryos.
Extended Data Figure 3 Barcode generation probabilities and number of Polylox recombination events in acutely labelled B cells.
a, Barcode generation probabilities were computed for a set of barcodes found experimentally (mouse 3, Extended Data Table 1, n = 506 barcodes) with and without length dependence of recombination rate, as described in Supplementary Methods. b, To compare the frequencies of individual barcode segments (‘letters’) generated by the model with experimental data, we focused on data from a Rosa26Polylox/CreERT2 mouse treated with tamoxifen (Fig. 3a, mouse 1), from which about 15,000 acutely barcoded B cells were analysed. To simulate barcode generation in 15,000 cells, 15,000 barcodes were drawn (with the frequencies of recombination events shown in e). This procedure was repeated 500 times to obtain standard deviations. c, Measured and computed distributions of fragment lengths (experimental data and simulations as in b). d, The observed and measured distributions of the 180 possible pairs of adjacent segments (experimental data and simulations as in b); the unrecombined pairs are particularly abundant. The PacBio instrument loads longer fragments less efficiently than shorter ones. Because of this bias, we restricted the analysis in b–d to fragments with 1, 3 and 5 segments. e, For all barcodes found in B cells from the mouse in b, we computed the minimal number of recombination events (excisions or/and inversions) needed to generate the barcode. All barcodes can be generated with six or fewer recombination events. The cumulative distribution of event frequencies is shown. Similar distributions were obtained in the reported barcoding experiments with Rosa26PolyloxTie2MCM mice. f, Barcodes generated once or multiple times in a simulated sample of 15,000 cells (as described in b). On average, 2,920 ± 35 (mean ± s.d.) different barcodes were generated with 15,000 draws. g, Measured barcode frequencies versus computed generation probabilities. For all barcodes retrieved in adult mice after barcode induction in embryonic HSC progenitors (Fig. 4, Exp. 1 and 2), we binned total barcode frequencies according to generation probabilities and calculated boxplot statistics of observed barcode frequencies for each bin (red line, median; box ends, 25% and 75% percentiles; bars, most extreme data points not considered outliers; red dots, outliers, n = 4, 95, 175, 97, 73, 28, 11 barcodes (left) and n = 4, 102, 180, 75, 42, 31, 14 barcodes (right)). From generation probabilities of 0.1 to 10−4, observed median frequency and probability of generation were overall correlated, showing that barcodes generated with higher probability are recovered more frequently. By contrast, for barcodes with generation probabilities <10−4, their median frequency of observation was independent of the probability of generation, indicating that these barcodes have each been generated in the smallest possible unit, a single embryonic HSC progenitor.
Extended Data Figure 4 Histogram of apparent HSC clone sizes.
For the single HSC sequencing data (Fig. 4, Exp. 1 and 2) we show separately the histogram of apparent clone sizes. An apparent clone is defined by all HSCs that contain the same barcode. These apparent clones are unlikely to all be biological clones, owing to the inclusion of abundant barcodes that may have been generated in more than one embryonic HSC progenitor. For the analysis of rare barcodes that are highly likely to define true clones generated from single HSC progenitors, see Fig. 4b, e.
Extended Data Figure 5 Overview of FACS gating strategies.
a–k, Distinctive surface marker combinations applied for the isolation of specific cell populations are depicted. Pre-gated lineage markers are indicated above the first plot of each panel. Not shown is the additional gating of all populations for size (FSC, SSC) and viability (Sytox blue−). For the complete listing of antibodies and marker phenotypes, see also Methods. a, Isolation of Kit+ Sca1+ stem cells (HSCs and ST-HSCs) and multipotent progenitors (MPPs), upper right, and Kit+ Sca1– myeloid progenitors (CMPs and GMPs), lower right, from bone marrow. b, Characterization of bone marrow CLPs. c, Definition of pre-B cells (Fr. B and Fr. C) in bone marrow. d, Thymic pre-T cells (DN2 and DN3). e, Gating of nucleated erythrocyte progenitors in the bone marrow (EryP II–IV, upper right, and EryP I, lower right). f, CD4 or CD8 single-positive T cells from spleen. g, Classical CD19+ splenic B cells. h, Neutrophilic granulocytes from the spleen. i, Splenic monocytes. j, Non-classical B cells (B1a and B1b) from the peritoneal cavity. k, Classical CD19+ B2 cells from peritoneal cavity.
Extended Data Figure 6 Adult barcode distributions in embryonically induced mice.
Mice as in Fig. 4a, Exp. 2. a, Heatmap of all barcodes found in HSCs (first lane) and the indicated erythroid, myeloid and lymphoid lineages in Exp. 2. b, Heatmap of peripheral barcodes (Pgen < 10−4, and detected in two independent samples of the same population) sorted according to lineage output in Exp. 2. Frequencies of barcodes are represented by colour-coded scales on the right.
Extended Data Figure 7 Clustering of cell types according to all mutual correlations reveals robust dichotomy between common myelo-erythroid and common lymphoid development.
All data from adult mice with embryonically induced barcodes (Fig. 4a, Exp. 2). a–c, Barcode frequencies in CD8 T cells versus B lymphocytes (B2) (a), in granulocytes (Gr) versus B lymphocytes (B2) (b) and granulocytes (Gr) versus CD4 T cells (c). Data in a–c are from Exp. 1, and each dot is an individual rare barcode with n = 48 (a), n = 49 (b) and n = 53 (c). d, Hierarchical clustering (with distance 1 – Spearman rank correlation coefficient, as described in Fig. 5d) applied to rare and reliably sampled barcodes found in indicated populations in Exp. 2 (n = 50). e, Hierarchical clustering as described in d but applied to all barcodes found in peripheral cells in Exp. 1 (n = 506) and Exp. 2 (n = 496). The inclusion of redundant barcodes reduces differences in correlations, but the split between common myelo-erythroid and common lymphoid is evident. f, Clustering as described in d but applied to rare multilineage barcodes (found in at least one erythroid, granulocyte and lymphocyte population, analogous to Fig. 4f; Exp.1, n = 16 and Exp. 2, n = 25). g, Clustering as described in d but applied only to barcodes found in adult HSCs, including redundant ones (shown in Fig. 4d; Exp.1, n = 54 and Exp. 2, n = 56). h, Summary of Spearman rank correlations (mean and 95% confidence bounds computed by non-parametric bootstrap) of GMPs versus the indicated lineages (for CMPs, see Fig. 5h); rare barcodes are from Exp. 2, n = 30–44.
Extended Data Figure 8 Polylox barcoding of haematopoiesis in adult mice (all data from adult mice with barcodes induced as adults).
a, Barcodes were induced by tamoxifen treatment of adult Rosa26PolyloxTie2MCM mice, and the indicated cell populations were analysed at 11–13 months of age. b, Tamoxifen treatment of adult Rosa26PolyloxTie2MCM mice (Extended Data Table 1) induced Polylox recombination in HSCs and, to a lesser extent, also in downstream stem and progenitor cells, ST-HSCs, MPPs and CMPs (Supplementary Fig. 1e) c, Heatmaps of barcodes satisfying single-cell induction criteria (at the time of labelling) recovered in the indicated stem and progenitor cells, and mature cells in Exp. 3 (left) and Exp. 4 (right). Frequencies of barcodes are represented by colour code (left). d, Heatmaps for individual HSCs satisfying adult single-cell barcode induction criteria, and their lineage output in Exp. 3 (top) and Exp. 4 (bottom). Pgen for the multilineage barcodes were as follows: 1F92G45H3, 1.3 × 10−9; 123FG45, 2 × 10−5. e, The barcode overlap between two samples of the same cell population (granulocytes and CD4 T cells isolated from the peripheral blood; 30,000 cells per sample) was smaller than for embryonically labelled mice (Fig. 4c). f, Hierarchical clustering of rank correlations of barcodes from the indicated populations (Exp. 3, n = 129). The colour scale (not shown) for rank correlations is identical to the scale bar shown in Fig. 5i.
Extended Data Figure 9 Induction of Polylox recombination in tissues of all three germ layers.
a, To induce Polylox recombination in different tissues in vivo, we crossed the Rosa26Polylox allele into mice bearing the Rosa26CreERT2 allele, which encodes ubiquitously expressed, tamoxifen-regulated Cre, yielding Rosa26Polylox/CreERT2 mice. b, Adult Rosa26Polylox/CreERT2 mice were injected with tamoxifen or with oil only (vehicle control) according to the depicted schedule, and were analysed on day 5. c, Genomic DNA was prepared from indicated organs that represent developmental derivatives of all three germ layers: brain (ectoderm), muscle, spleen, and thymus (mesoderm), and liver and lung (endoderm). The Polylox cassette was amplified by PCR, and recombination in each tissue and for all time points was visualized by separating DNA fragments by gel electrophoresis (Supplementary Fig. 1f). The first lane is the PCR water control, the second lane is from Rosa26+/+ DNA template, and the third lane is from Rosa26Polylox(no Cre) template; all other lanes show data from Rosa26Polylox/CreERT2 mice for the indicated organs and conditions. The DNA sample and PCR result from the muscle oil control were not available.
Supplementary information
Supplementary Information
This file contains supplementary figure 1, data, tables 1-3 and methods. (PDF 6355 kb)
Rights and permissions
About this article
Cite this article
Pei, W., Feyerabend, T., Rössler, J. et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017). https://doi.org/10.1038/nature23653
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23653
This article is cited by
-
Made to order: emergency myelopoiesis and demand-adapted innate immune cell production
Nature Reviews Immunology (2024)
-
Extracting, filtering and simulating cellular barcodes using CellBarcode tools
Nature Computational Science (2024)
-
Deciphering cell states and genealogies of human haematopoiesis
Nature (2024)
-
In vivo clonal tracking reveals evidence of haemangioblast and haematomesoblast contribution to yolk sac haematopoiesis
Nature Communications (2023)
-
The transcriptional legacy of developmental stochasticity
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.