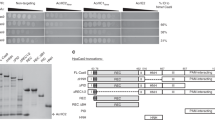
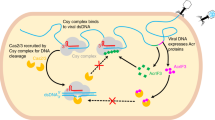
Abstract
CRISPR–Cas9 systems are bacterial adaptive immune systems that defend against infection by phages. Through the RNA-guided endonuclease activity of Cas9 they degrade double-stranded DNA with a protospacer adjacent motif (PAM) and sequences complementary to the guide RNA1,2,3,4,5. Recently, two anti-CRISPR proteins (AcrIIA2 and AcrIIA4 from Listeria monocytogenes prophages) were identified, both of which inhibit Streptococcus pyogenes Cas9 (SpyCas9) and L. monocytogenes Cas9 activity in bacteria and human cells6. However, the mechanism of AcrIIA2- or AcrIIA4-mediated Cas9 inhibition remains unknown. Here we report a crystal structure of SpyCas9 in complex with a single-guide RNA (sgRNA) and AcrIIA4. Our data show that AcrIIA2 and AcrIIA4 interact with SpyCas9 in a sgRNA-dependent manner. The structure reveals that AcrIIA4 inhibits SpyCas9 activity by structurally mimicking the PAM to occupy the PAM-interacting site in the PAM-interacting domain, thereby blocking recognition of double-stranded DNA substrates by SpyCas9. AcrIIA4 further inhibits the endonuclease activity of SpyCas9 by shielding its RuvC active site. Structural comparison reveals that formation of the AcrIIA4-binding site of SpyCas9 is induced by sgRNA binding. Our study reveals the mechanism of SpyCas9 inhibition by AcrIIA4, providing a structural basis for developing ‘off-switch’ tools for SpyCas9 to avoid unwanted genome edits within cells and tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Makarova, K. S. et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015)
Wiedenheft, B., Sternberg, S. H. & Doudna, J. A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 (2012)
Marraffini, L. A. CRISPR–Cas immunity in prokaryotes. Nature 526, 55–61 (2015)
Westra, E. R. et al. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46, 311–339 (2012)
Sorek, R., Lawrence, C. M. & Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266 (2013)
Rauch, B. J. et al. Inhibition of CRISPR–Cas9 with bacteriophage proteins. Cell 168, 150–158.e10 (2017)
Barrangou, R. & Marraffini, L. A. CRISPR–Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244 (2014)
Horvath, P. & Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 (2010)
van der Oost, J., Jore, M. M., Westra, E. R., Lundgren, M. & Brouns, S. J. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34, 401–407 (2009)
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012)
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012)
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014)
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014)
Jinek, M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014)
Jiang, F., Zhou, K., Ma, L., Gressel, S. & Doudna, J. A. Structural biology. A Cas9–guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 (2015)
Jiang, W. & Marraffini, L. A. CRISPR–Cas: new tools for genetic manipulations from bacterial immunity systems. Annu. Rev. Microbiol. 69, 209–228 (2015)
Sternberg, S. H. & Doudna, J. A. Expanding the biologist’s toolkit with CRISPR–Cas9. Mol. Cell 58, 568–574 (2015)
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157, 1262–1278 (2014)
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M. & Joung, J. K. Improving CRISPR–Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284 (2014)
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013)
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013)
Yen, S. T. et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev. Biol. 393, 3–9 (2014)
Orthwein, A. et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015)
Wright, A. V. et al. Rational design of a split-Cas9 enzyme complex. Proc. Natl Acad. Sci. USA 112, 2984–2989 (2015)
Nihongaki, Y., Kawano, F., Nakajima, T. & Sato, M. Photoactivatable CRISPR–Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760 (2015)
Nuñez, J. K., Harrington, L. B. & Doudna, J. A. Chemical and biophysical modulation of Cas9 for tunable genome engineering. ACS Chem. Biol. 11, 681–688 (2016)
Pawluk, A. et al. Naturally occurring off-switches for CRISPR–Cas9. Cell 167, 1829–1838.e9 (2016)
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015)
Chowdhury, S. et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 169, 47–57.e11 (2017)
Jorgensen, I., Rayamajhi, M. & Miao, E. A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164 (2017)
Otwinowski, Z. M. W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D . et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
DeLano, W. L. PyMOL Molecular Viewer (http://www.pymol.org) (2002)
Acknowledgements
We thank J. He at Shanghai Synchrotron Radiation Facility (SSRF) for help with data collection. We thank J. Chai for critical reading of the manuscript. This research was funded by the National Natural Science Foundation of China grant no. 31422014, 31450001 and 31300605 to Z.H.
Author information
Authors and Affiliations
Contributions
D.D., M.G. and S.W. expressed, purified, characterized and crystallized the AcrIIA4–SpyCas9–sgRNA complex with the help of S.W., Z.X., J.Y. and Z.X. D.D., M.G., Y.Z. and Z.H. carried out crystallographic studies. D.D., M.G. and Z.H. prepared the figures. D.D. and M.G. performed in vitro transcription of sgRNA, in vitro dsDNA cleavage, GST pull-down, gel filtration, microscale thermophoresis and electrophoretic mobility shift assay experiments with the help of S.W., Z.X., J.Y. and Z.X. Z.H., D.D., M.G. and S.W. wrote the paper. All authors contributed to the manuscript preparation. Z.H. designed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks S. Bailey, J. van der Oost and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 AcrIIA2 and AcrIIA4 specifically interact with sgRNA-bound SpyCas9.
Binding assays were carried out between the anti-CRISPR protein of AcrIIA2 or AcrIIA4 and the GST-tagged CRISPR protein of SpyCas9 or NmeCas9 in the presence or absence of cognate sgRNA. The purified GST–SpyCas9 or GST–NmeCas9 protein was first bound to glutathione sepharose beads in the presence or absence of sgRNA, and then the beads were incubated them with AcrIIA2 or AcrIIA4 protein. After extensive washing, the bound proteins were visualized by Coomassie staining following SDS–PAGE. Data shown are the representative of three replicates.
Extended Data Figure 2 Alignment of Cas9 protein sequences.
Multiple sequence alignment of the amino acid sequences of type II-A Cas9 proteins from Streptococcus pyogenes (GI 15675041), Listeria monocytogenes J0161 (GI 345535315), Listeria innocua Clip11262 (GI 16414891), and type II-C Cas9 proteins of Neisseria meningitidis (GI 518572566), Pasteurella multocida subsp. multocida str. Pm70 (GI 218767588), aligned using MUSCLE. Residues with more than 70% similarity are shown in red and boxed in blue. Residues involved in interaction with AcrIIA4 are indicated.
Extended Data Figure 3 Structural comparison of SpyCas9–sgRNA–DNA and SpyCas9–sgRNA–AcrIIA4.
a, Structural superimposition of SpyCas9–sgRNA–DNA (PDB code, 4UN3) and SpyCas9–sgRNA–AcrIIA4. b, GST pull-down assays to verify the structural determinants for preferential binding of AcrIIA4 to SpyCas9. Wild-type or mutant GST-fused AcrIIA4 proteins were first bound to glutathione sepharose beads and incubated with sgRNA-preloaded SpyCas9 (or mutant) protein as indicated. After extensive washing, the bound proteins were visualized by Coomassie staining following SDS–PAGE. c, Enzymatic activity assays to verify structural determinants for specific AcrIIA4–SpyCas9 interaction. The assays were performed as described in Fig. 1b. Data shown are representative of three independent experiments. d, AcrIIA2 and AcrIIA4 compete with PAM-containing dsDNA for binding to the SpyCas9–sgRNA. sgRNA-preloaded GST–SpyCas9 protein was first mixed with AcrIIA2 or AcrIIA4 at 4 °C and incubated for 15 min, followed by addition of PAM-containing dsDNA into the reaction mixtures. After 15 min incubation, the reactions were stopped by adding loading buffer for denaturing gel and the reaction mixtures were loaded onto glutathione sepharose beads and incubated for 15 min. After extensive washing, the bound proteins were visualized by Coomassie staining following SDS–PAGE. Data shown are representative of three independent experiments. e, Electrophoretic mobility shift assay results showing that AcrIIA2 and AcrIIA4 compete with PAM-containing dsDNA for binding to the SpyCas9–sgRNA. The fluorophore-labelled dsDNA and AcrIIA2 (upper panel) or AcrIIA4 (lower panel) were added to sgRNA-preloaded inactive SpyCas9(D10A/H840A) simultaneously. Molar ratios of SpyCas9–anti-CRISPR protein are shown at the top of each lane. The reaction mixtures were run on 6% native polyacrylamide gels and visualized by fluorescence imaging (800 nm). Data shown are representative of three independent experiments.
Extended Data Figure 4 Structural comparison of SpyCas9–sgRNA and SpyCas9.
Structural superimposition of SpyCas9–sgRNA (PDB code, 4ZT0) and SpyCas9 (PDB code, 4CMP). The interface of AcrIIA4 and SpyCas9 is circled in black dashed line.
Supplementary information
Supplementary Figure
This file contains the uncropped gels. (PDF 959 kb)
Source data
Rights and permissions
About this article
Cite this article
Dong, D., Guo, M., Wang, S. et al. Structural basis of CRISPR–SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436–439 (2017). https://doi.org/10.1038/nature22377
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22377
This article is cited by
-
An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding
Nature Communications (2024)
-
Inhibitors of bacterial immune systems: discovery, mechanisms and applications
Nature Reviews Genetics (2024)
-
Anti-CRISPR proteins: a weapon of phage-bacterial arm race for genome editing
The Nucleus (2023)
-
A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1
Nature Communications (2022)
-
A general approach to identify cell-permeable and synthetic anti-CRISPR small molecules
Nature Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.