Abstract
Chromatin and metabolic states both influence lifespan, but how they interact in lifespan regulation is largely unknown. The COMPASS chromatin complex, which trimethylates lysine 4 on histone H3 (H3K4me3), regulates lifespan in Caenorhabditis elegans. However, the mechanism by which H3K4me3 modifiers affect longevity, and whether this mechanism involves metabolic changes, remain unclear. Here we show that a deficiency in H3K4me3 methyltransferase, which extends lifespan, promotes fat accumulation in worms with a specific enrichment of mono-unsaturated fatty acids (MUFAs). This fat metabolism switch in H3K4me3 methyltransferase-deficient worms is mediated at least in part by the downregulation of germline targets, including S6 kinase, and by the activation of an intestinal transcriptional network that upregulates delta-9 fatty acid desaturases. Notably, the accumulation of MUFAs is necessary for the lifespan extension of H3K4me3 methyltransferase-deficient worms, and dietary MUFAs are sufficient to extend lifespan. Given the conservation of lipid metabolism, dietary or endogenous MUFAs could extend lifespan and healthspan in other species, including mammals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Benayoun, B. A., Pollina, E. A. & Brunet, A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 16, 593–610 (2015)
Sen, P., Shah, P. P., Nativio, R. & Berger, S. L. Epigenetic mechanisms of longevity and aging. Cell 166, 822–839 (2016)
Miller, M. et al. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation 123, 2292–2333 (2011)
Hansen, M., Flatt, T. & Aguilaniu, H. Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 17, 10–19 (2013)
Shilatifard, A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95 (2012)
Greer, E. L. et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466, 383–387 (2010)
O’Rourke, E. J., Soukas, A. A., Carr, C. E. & Ruvkun, G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 10, 430–435 (2009)
Brooks, K. K., Liang, B. & Watts, J. L. The influence of bacterial diet on fat storage in C. elegans. PLoS One 4, e7545 (2009)
Espelt, M. V., Estevez, A. Y., Yin, X. & Strange, K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. J. Gen. Physiol. 126, 379–392 (2005)
Marré, J ., Traver, E. C. & Jose, A. M. Extracellular RNA is transported from one generation to the next in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 113, 12496–12501 (2016)
Kumsta, C. & Hansen, M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One 7, e35428 (2012)
Watts, J. L. & Browse, J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 272, 263–269 (2000)
Peyou-Ndi, M. M., Watts, J. L. & Browse, J. Identification and characterization of an animal delta(12) fatty acid desaturase gene by heterologous expression in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 376, 399–408 (2000)
Robert, V. J. et al. The SET-2/SET1 histone H3K4 methyltransferase maintains pluripotency in the Caenorhabditis elegans germline. Cell Reports 9, 443–450 (2014)
Pferdehirt, R. R., Kruesi, W. S. & Meyer, B. J. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 25, 499–515 (2011)
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012)
Kapahi, P. et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465 (2010)
Shi, X. et al. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 54, 2504–2514 (2013)
Yang, F. et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 (2006)
Taubert, S., Van Gilst, M. R., Hansen, M. & Yamamoto, K. R. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 20, 1137–1149 (2006)
Van Gilst, M. R., Hadjivassiliou, H., Jolly, A. & Yamamoto, K. R. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3, e53 (2005)
Brock, T. J., Browse, J. & Watts, J. L. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2, e108 (2006)
Brock, T. J., Browse, J. & Watts, J. L. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics 176, 865–875 (2007)
Shmookler Reis, R. J. et al. Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany, N.Y.) 3, 125–147 (2011)
Goudeau, J. et al. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 9, e1000599 (2011)
O’Rourke, E. J., Kuballa, P., Xavier, R. & Ruvkun, G. ω-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 27, 429–440 (2013)
Ratnappan, R. et al. Germline signals deploy NHR-49 to modulate fatty-acid β-oxidation and desaturation in somatic tissues of C. elegans. PLoS Genet. 10, e1004829 (2014)
Lee, D. et al. SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat. Genes Dev. 29, 2490–2503 (2015)
Magnuson, K., Jackowski, S., Rock, C. O. & Cronan, J. E., Jr. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57, 522–542 (1993)
Gillingham, L. G., Harris-Janz, S. & Jones, P. J. Dietary monounsaturated fatty acids are protective against metabolic syndrome and cardiovascular disease risk factors. Lipids 46, 209–228 (2011)
Sijen, T. et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 (2001)
Greer, E. L. et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 (2007)
Pino, E. C., Webster, C. M., Carr, C. E. & Soukas, A. A. Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J. Vis. Exp. 73, 50180 (2013)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Reinke, V., Gil, I. S., Ward, S. & Kazmer, K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311–323 (2004)
Wang, X. et al. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10, 213 (2009)
Gerstein, M. B. et al. Comparative analysis of the transcriptome across distant species. Nature 512, 445–448 (2014)
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009)
Deline, M. L., Vrablik, T. L. & Watts, J. L. Dietary supplementation of polyunsaturated fatty acids in Caenorhabditis elegans. J. Vis. Exp. 81, 50879 (2013)
Acknowledgements
We thank A. Jose, A. Rechtsteiner, S. Strome and R. Waterston for sharing expression data and strains pre-publication; A. Fire, M. Hansen, S. Kim, F. Palladino, D. Pattabiraman, Y. Zhang and the Caenorhabditis Genetics Center for plasmids and strains; M. Hansen, E. O’Rourke, L. Booth, C-K. Hu, D. Leeman, J. Lim and S. Mahmoudi for reading the manuscript; A. Fire, O. Gozani, S. Kim and Brunet laboratory members for discussions; A. Chien for GC–MS consulting; and J. Coller at the Stanford Functional Genomics Facility. Supported by NIH DP1AG044848 (A.B.), NIH R01AG054201 (A.B. and W.B.M.), NIH R01AG044346 (W.B.M.), a Stanford Mass Spectrometry grant (S.H. and A.B.), NSF Graduate Research Fellowship, Stanford Graduate Fellowship and NIH T32AG047126 (S.H.), and NIH T32AG047126 and NIH F32AG051337 (E.A.S.).
Author information
Authors and Affiliations
Contributions
S.H. conceived the study under the guidance of A.B. S.H. performed all the experiments except those specified below. E.A.S. planned and performed Nile Red and FAT-5/FAT-7 reporter imaging, time-course RT–qPCR and one oleic acid supplementation lifespan experiment. C.G.S.-G. planned and performed SBP-1 localization experiments and generated set-2 transgenic lines under the guidance of W.B.M. K.H. analysed the RNA-seq data. S.H. wrote the paper with the help of A.B. and E.A.S. C.G.S.-G., K.H. and W.B.M. provided feedback.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks H. Aguilaniu, M. Kaeberlein and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
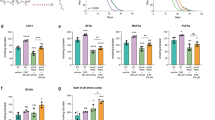
Extended Data Figure 1 Deficiency in H3K4me3 modifiers leads to fat accumulation in the intestine without altering fertility.
a, ORO quantification. Mean ± s.d., n ≥ 17 worms per condition. b, ORO images. Scale bars, 100 μm. c, ORO quantification. Mean ± s.d., n ≥ 9 dissected intestines per condition. d, e, Nile Red staining and quantification. Mean ± s.d., n ≥ 7 dissected intestines (d) or n ≥ 11 worms (e) per condition. Scale bars, 100 μm. f, Autofluorescence and quantification. Mean ± s.d., n ≥ 17 worms per condition. Scale bars, 100 μm. g, Fertility quantification of live brood size (i), fertilized eggs (ii), and unfertilized oocytes (iii) laid per worm. Mean ± s.d., n ≥ 25 worms per condition. h, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with two or three biological replicates. i, RT–qPCR. Mean ± s.e.m. from two independent experiments, each with two or three biological replicates. j, ORO quantification. Mean ± s.d., n ≥ 26 worms per condition. k, ORO quantification. Mean ± s.d., n ≥ 29 worms per condition. l, RT–qPCR. Mean ± s.e.m. of three biological replicates. m, Differential interference contrast (DIC) (Nomarski) and GFP fluorescence images. n, ORO quantification. Mean ± s.d., n ≥ 26 worms per condition. a–f, k (rescue line number 1), n, Representative of two experiments. P values: a, c–g, two-tailed Mann–Whitney; h, i, two-tailed Mann–Whitney with Benjamini–Hochberg correction; j, k, n, Kruskal–Wallis with Dunn’s correction; l, two-tailed t-test with Benjamini–Hochberg correction. *P < 0.05, **P < 0.01.
Extended Data Figure 2 The delta-9 desaturases FAT-5 and FAT-7 support MUFA accumulation in H3K4me3 methyltransferase-deficient worms.
a, GC–MS quantification of MUFAs. Mean ± s.e.m. of two independent experiments, each with three biological replicates. b, c, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with three biological replicates. d, GC–MS quantification of MUFAs. Mean ± s.e.m. of two independent experiments, each with two or three biological replicates. e, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with three biological replicates. P values: a–e, two-tailed Mann–Whitney with Benjamini–Hochberg correction. *P < 0.05, **P < 0.01.
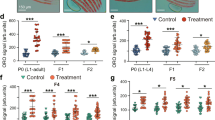
Extended Data Figure 3 RNA-seq on micro-dissected germlines and intestines and functional validation of ASH-2 targets.
a, RNA-seq tissue sample collection pipeline. b, Principal component analysis (PCA) with both intestinal and germline samples (left), only intestinal samples (middle) or only germline samples (right). c, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with two or three biological replicates. d – g, ORO quantification. Mean ± s.d., n ≥ 15 worms per condition. h, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with three biological replicates. i, j, ORO quantification. Mean ± s.d., n ≥ 21 (i) and n ≥ 27 (j) worms per condition. k, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with three biological replicates. l, GC–MS. Mean ± s.e.m. of two independent experiments, each with three biological replicates. m, ORO quantification. Mean ± s.d., n ≥ 19 worms per condition. n, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with three biological replicates. o, Lifespan extension by ash-2 RNAi is reduced in rsks-1 mutants (13.12%) compared to wild-type worms (29.20%) (P < 0.0001, two-way ANOVA). p, let-363 RNAi and daf-15 RNAi extend lifespan in wild-type worms (P < 0.0001, log-rank), but not in set-2 mutants. d (except rrf-1 data), i, j, Representative of two experiments. P values: c, k, l, n, two-tailed Mann–Whitney with Benjamini–Hochberg correction; h, two-tailed Mann-Whitney; d–g, i, j, m, Kruskal–Wallis with Dunn’s correction. *P < 0.05, **P < 0.01.
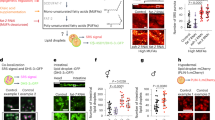
Extended Data Figure 4 Role of SBP-1, MDT-15, NHR-49 and NHR-80 in the fat accumulation and longevity of H3K4me3 methyltransferase-deficient worms.
a–d, Images and quantification of SBP-1 nuclear accumulation. Mean ± s.d. of two independent experiments, each with 4–6 nuclei per worm of ≥ 8 worms per condition. e, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with three biological replicates. f, GC–MS quantification of MUFAs. Mean ± s.e.m. of two independent experiments, each with two or three biological replicates. g, ORO quantification. Mean ± s.d., n ≥ 21 worms per condition. h, ash-2 RNAi extends lifespan in wild-type worms (P < 0.0001, log-rank), but not in mdt-15 RNAi worms. i, ORO quantification, Mean ± s.d., n ≥ 24 worms per condition. j, ash-2 RNAi extends lifespan in both wild-type worms and nhr-49 mutants (P < 0.0001, log-rank). k, ORO quantification. Mean ± s.d., n ≥ 27 worms per condition. l, ash-2 RNAi extends lifespan in both wild-type worms and nhr-80 mutants (P < 0.0001, log-rank). i–k, Representative of two experiments. P values: a, two-tailed Mann–Whitney; b–d, g, i, k, Kruskal–Wallis with Dunn’s correction; e, f, two-tailed Mann–Whitney with Benjamini–Hochberg correction. *P < 0.05.
Extended Data Figure 5 Delta-9 desaturases FAT-6 and FAT-7 and MUFA oleic acid mediate the longevity of H3K4me3 methyltransferase-deficient worms.
a, ORO quantification. Mean ± s.d., n ≥ 29 worms per condition. b, ash-2 RNAi leads to lifespan extension in control (P < 0.0001, log-rank), but not in fat-6 and fat-7 double mutants. c, ash-2 RNAi extends lifespan in both wild-type worms and fat-7 single mutants (P < 0.0001, log-rank). d, ash-2 RNAi extends lifespan in fat-6 single mutants (P = 0.0162, log-rank), but lifespan extension by ash-2 RNAi is reduced in fat-6 mutants (9.26%) compared to wild-type worms (20.46%) (P = 0.0072, two-way ANOVA). e, ash-2 RNAi extends lifespan in fat-5 and fat-7 double mutants (P < 0.0001, log-rank). f, ash-2 RNAi extends lifespan in fat-5 and fat-6 double mutants (P = 0.0002, log-rank), but lifespan extension by ash-2 RNAi is reduced in fat-5 and fat-6 double mutants (14.03%) compared to wild-type worms (20.46%) (P = 0.0358, two-way ANOVA). g, h, GC–MS quantification of oleic acid. Mean ± s.e.m. of three biological replicates. i, ORO quantification. Mean ± s.d., n ≥ 13 worms per condition. Boxed conditions are identical to Fig. 4g. j, Oleic acid supplementation extends lifespan (P < 0.0001, log-rank), which is not further extended by ash-2 RNAi. Oleic acid supplementation extends lifespan in ash-2 and fat-7 double RNAi worms (P < 0.0001, log-rank). Boxed conditions are identical to Fig. 4h. k, ORO quantification. Mean ± s.d., n ≥ 21 worms per condition. l, GC–MS quantification of MUFAs. Mean ± s.e.m. of two independent experiments, each with two or three biological replicates. m, RT–qPCR. Mean ± s.e.m. of two independent experiments, each with three biological replicates. n, ORO quantification. Mean ± s.d., n ≥ 25 worms per condition. o, ash-2 RNAi extends lifespan in control (P < 0.0001, log-rank) but not fat-2 RNAi worms. c, i, j, n, o, Representative of two experiments. P values: a, g–i, k, n, Kruskal–Wallis with Dunn’s correction; l, m, two-tailed Mann–Whitney with Benjamini–Hochberg correction. *P < 0.05, **P < 0.01.
Extended Data Figure 6 Dietary supplementation with MUFAs, but not PUFAs, extends lifespan in wild-type worms.
a, GC–MS. Mean ± s.e.m. of two independent experiments, each with two or three biological replicates. b, Cis-vaccenic acid supplementation extends lifespan in wild-type worms (P < 0.0001, log-rank). Inset: ORO quantification. Mean ± s.d., n ≥ 20 worms per condition. c, GC–MS. Mean ± s.e.m. of two independent experiments, each with three biological replicates. d, Linoleic acid supplementation does not extend lifespan in wild-type worms. Inset: ORO quantification. Mean ± s.d., n ≥ 23 worms per condition. e, GC–MS. Mean ± s.e.m. of two independent experiments, each with three biological replicates. f, Alpha-linolenic acid supplementation does not increase the lifespan of wild-type worms. Inset: ORO quantification. Mean ± s.d., n ≥ 23 worms per condition. g, RT–qPCR. Mean ± s.e.m. of three biological replicates. h, ORO quantification. Mean ± s.d., n ≥ 54 worms per condition. i, Overexpression of FAT-7 extends lifespan (P < 0.0001, log-rank), but this lifespan is not extended further by dietary oleic acid. Boxed regions are identical to Fig. 5g. j, Proposed model by which ash-2 deficiency in the germline could lead to the fat metabolic switch in the intestine. b, d, f, Representative of two experiments. P values: a, c, e, two-tailed Mann–Whitney with Benjamini–Hochberg correction; b inset, d inset, f inset, two-tailed Mann–Whitney; g, Kruskal–Wallis test with Dunn’s correction (non-significant, probably owing to small sample size). One-way ANOVA with Bonferroni’s correction. h, Kruskal–Wallis with Dunn’s correction; *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-8 and a Supplementary Table guide. (ZIP 3003 kb)
Rights and permissions
About this article
Cite this article
Han, S., Schroeder, E., Silva-García, C. et al. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544, 185–190 (2017). https://doi.org/10.1038/nature21686
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21686
This article is cited by
-
Evolutionarily related host and microbial pathways regulate fat desaturation in C. elegans
Nature Communications (2024)
-
The chromatin factors SET-26 and HCF-1 oppose the histone deacetylase HDA-1 in longevity and gene regulation in C. elegans
Nature Communications (2024)
-
Reducing the metabolic burden of rRNA synthesis promotes healthy longevity in Caenorhabditis elegans
Nature Communications (2024)
-
Fatty acid- and retinol-binding protein 6 does not control worm fatty acid content in Caenorhabditis elegans but might play a role in Haemonchus contortus parasitism
Parasites & Vectors (2023)
-
Early life changes in histone landscape protect against age-associated amyloid toxicities through HSF-1-dependent regulation of lipid metabolism
Nature Aging (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.