Abstract
Since its discovery in 1989, efforts to grow clinical isolates of the hepatitis C virus (HCV) in cell culture have met with limited success. Only the JFH-1 isolate has the capacity to replicate efficiently in cultured hepatoma cells without cell culture-adaptive mutations1,2,3. We hypothesized that cultured cells lack one or more factors required for the replication of clinical isolates. To identify the missing factors, we transduced Huh-7.5 human hepatoma cells with a pooled lentivirus-based human complementary DNA (cDNA) library, transfected the cells with HCV subgenomic replicons lacking adaptive mutations, and selected for stable replicon colonies. This led to the identification of a single cDNA, SEC14L2, that enabled RNA replication of diverse HCV genotypes in several hepatoma cell lines. This effect was dose-dependent, and required the continuous presence of SEC14L2. Full-length HCV genomes also replicated and produced low levels of infectious virus. Remarkably, SEC14L2-expressing Huh-7.5 cells also supported HCV replication following inoculation with patient sera. Mechanistic studies suggest that SEC14L2 promotes HCV infection by enhancing vitamin E-mediated protection against lipid peroxidation. This provides a foundation for development of in vitro replication systems for all HCV isolates, creating a useful platform to dissect the mechanisms by which cell culture-adaptive mutations act.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Blight, K. J., Kolykhalov, A. A. & Rice, C. M. Efficient initiation of HCV RNA replication in cell culture. Science 290, 1972–1974 (2000)
Saeed, M. et al. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob. Agents Chemother. 56, 5365–5373 (2012)
Kato, T . et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125, 1808–1817 (2003)
Mohd Hanafiah, K., Groeger, J., Flaxman, A. D. & Wiersma, S. T. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57, 1333–1342 (2013)
Gottwein, J. M. et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J. Virol. 84, 5277–5293 (2010)
Murayama, A. et al. The NS3 helicase and NS5B-to-3′X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells. J. Virol. 81, 8030–8040 (2007)
Murayama, A. et al. RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells. PLoS Pathog. 6, e1000885 (2010)
Bukh, J. et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl Acad. Sci. USA 99, 14416–14421 (2002)
Allen-Baume, V., Segui, B. & Cockcroft, S. Current thoughts on the phosphatidylinositol transfer protein family. FEBS Lett. 531, 74–80 (2002)
Aravind, L., Neuwald, A. F. & Ponting, C. P. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr. Biol. 9, R195–R197 (1999)
Kempná, P. et al. Cloning of novel human SEC14p-like proteins: ligand binding and functional properties. Free Radic. Biol. Med. 34, 1458–1472 (2003)
Jones, C. T. et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nature Biotechnol. 28, 167–171 (2010)
Farci, P. et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl Acad. Sci. USA 93, 15394–15399 (1996)
Washburn, M. L. et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology 140, 1334–1344 (2011)
Panagabko, C. et al. Ligand specificity in the CRAL-TRIO protein family. Biochemistry 42, 6467–6474 (2003)
Ni, J. et al. Tocopherol-associated protein suppresses prostate cancer cell growth by inhibition of the phosphoinositide 3-kinase pathway. Cancer Res. 65, 9807–9816 (2005)
Mokashi, V., Singh, D. K. & Porter, T. D. Supernatant protein factor stimulates HMG-CoA reductase in cell culture and in vitro . Arch. Biochem. Biophys. 433, 474–480 (2005)
Mokashi, V. & Porter, T. D. Supernatant protein factor requires phosphorylation and interaction with Golgi to stimulate cholesterol synthesis in hepatoma cells. Arch. Biochem. Biophys. 435, 175–181 (2005)
Neuzil, J., Dong, L. F., Wang, X. F. & Zingg, J. M. Tocopherol-associated protein-1 accelerates apoptosis induced by alpha-tocopheryl succinate in mesothelioma cells. Biochem. Biophys. Res. Commun. 343, 1113–1117 (2006)
Yamane, D. et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nature Med. 20, 927–935 (2014)
Yoshimoto, T. et al. Positive modulation of IL-12 signaling by sphingosine kinase 2 associating with the IL-12 receptor beta 1 cytoplasmic region. J. Immunol. 171, 1352–1359 (2003)
Poveda, E. et al. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res. 108, 181–191 (2014)
Lohmann, V. et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 (1999)
Wose Kinge, C. N. et al. Hepatitis C virus genotype 5a subgenomic replicons for evaluation of direct-acting antiviral agents. Antimicrob. Agents Chemother. 58, 5386–5394 (2014)
Liehl, P. et al. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nature Med. 20, 47–53 (2014)
Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262, 250–263 (1999)
Lohmann, V., Korner, F., Dobierzewska, A. & Bartenschlager, R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75, 1437–1449 (2001)
Blight, K. J., McKeating, J. A., Marcotrigiano, J. & Rice, C. M. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77, 3181–3190 (2003)
Shimakami, T. et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140, 667–675 (2011)
Wakita, T. et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Med. 11, 791–796 (2005)
Lindenbach, B. D. et al. Complete replication of hepatitis C virus in cell culture. Science 309, 623–626 (2005)
Marukian, S. et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48, 1843–1850 (2008)
Fukuhara, T. et al. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J. Virol. 86, 7918–7933 (2012)
Rodriguez-Barrueco, R., Marshall, N. & Silva, J. M. Pooled shRNA screenings: experimental approach. Methods Mol. Biol. 980, 353–370 (2013)
Yi, Z. et al. Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS Pathog. 7, e1001255 (2011)
Schoggins, J. W. et al. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro . Proc. Natl Acad. Sci. USA 109, 14610–14615 (2012)
Suthar, M. S., Shabman, R., Madric, K., Lambeth, C. & Heise, M. T. Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. J. Virol. 79, 4219–4228 (2005)
Shabman, R. S. et al. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J. Virol. 81, 237–247 (2007)
Bredenbeek, P. J. et al. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84, 1261–1268 (2003)
Kinney, R. M. et al. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230, 300–308 (1997)
Chung, H. Y., Gu, M., Buehler, E., MacDonald, M. R. & Rice, C. M. Seed sequence-matched controls reveal limitations of small interfering RNA knockdown in functional and structural studies of hepatitis C virus NS5A-MOBKL1B interaction. J. Virol. 88, 11022–11033 (2014)
Cristea, I. M., Williams, R., Chait, B. T. & Rout, M. P. Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4, 1933–1941 (2005)
Pan, M. et al. Lipid peroxidation and oxidant stress regulate hepatic apolipoprotein B degradation and VLDL production. J. Clin. Invest. 113, 1277–1287 (2004)
Scheel, T. K. et al. Analysis of functional differences between hepatitis C virus NS5A of genotypes 1-7 in infectious cell culture systems. PLoS Pathog. 8, e1002696 (2012)
Maqbool, M. A. et al. Regulation of hepatitis C virus replication by nuclear translocation of nonstructural 5A protein and transcriptional activation of host genes. J. Virol. 87, 5523–5539 (2013)
Acknowledgements
We thank Y. Matsuura for HEP3B/miR-122 cells, J. Bukh for pJ6CF and pTNcc/GLuc, S. M. Lemon and D. Yamane for pH77S.3/GLuc, pH77S.3, and pH77D/GLuc, and D. Manor for [14C]RRR-α-tocopherol. We also thank E. Castillo and A. Webson for laboratory assistance, H. H. Hoffman and L. Andrus for technical input, and W. M. Schneider, M. M. Li and M. R. MacDonald for critical reading of the manuscript. This work was supported in part by the National Institutes of Health, NCI grant R01CA057973, NIAID grants R01AI072613 and R01AI099284 (to C.M.R.), NIDDK grant R01DK090317 and NIDA grant DA031095 (to A.D.B.), by a Helmsley Postdoctoral Fellowship for Basic and Translational Research on Disorders of the Digestive System at The Rockefeller University (to M.S.), and by a Liver Scholar Award from American Association for the Study of Liver Diseases (U.A.). The Greenberg Medical Research Institute, the Starr Foundation, the Ronald A. Shellow, M.D. Memorial Fund and anonymous donors provided the additional funding (to C.M.R.).
Author information
Authors and Affiliations
Contributions
M.S. and C.M.R. designed the project, analysed results, and wrote the manuscript. M.S., U.A., H.-Y.C. and C.E. performed the experimental work. A.D.B. and J.M.S. contributed reagents and advice.
Corresponding author
Ethics declarations
Competing interests
C.M.R. has equity in Apath, LLC, which holds commercial licenses for the Huh-7.5 cell line and certain HCV cell culture systems.
Extended data figures and tables
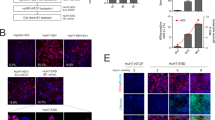
Extended Data Figure 1 Cured replicon cell clones supported replication of wild-type HCV.
Six of the 45 cell colonies obtained from cDNA screen and one control cell clone, S52 (adapted) that carried a cell culture-adapted HCV subgenomic replicon S52/SG-neo(AII), were treated for 15 days with a combination of an HCV NS5A inhibitor (daclatasvir, 1 nM) and an NS5B polymerase inhibitor (2′CMeA, 1 µM) to eliminate the replicating viral genomes. These cells were then transfected with the indicated wild-type HCV replicons and selected with G418 for 3 weeks. The resulting cell colonies were stained with crystal violet. Replication defective replicon, S52 GNN, with a catalytically inactive mutation in the NS5B polymerase served as a negative control.
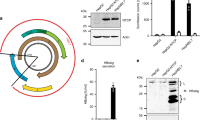
Extended Data Figure 2 The ectopic expression of SEC14L2 confers HCV permissiveness to human hepatoma cells.
a, b, SEC14L2 is not expressed in most cell lines. a, SEC14L2 mRNA levels were measured in the indicated cells by qPCR, and the values were normalized to those of the housekeeping gene, RPS11. Shown is the fold difference from fetal hepatocytes. The results are plotted as mean ± s.e.m. of two different cell stocks. b, SEC14L2 protein expression in the indicated cells was determined by immunoblotting with SEC14L2 rabbit polyclonal antibody. β-actin is included as a loading control. c, SEC14L2-expressing Huh-7.5 cells were transfected with the indicated wild-type subgenomic replicons. After 4 weeks of selection, HCV RNA was sequenced in six individual colonies from each of the replicons. HCV RNA levels in each of these colonies were determined by qRT–PCR. d, Hep3B/miR-122 and Huh-7 cells transduced with SEC14L2 or empty vector were transfected with in vitro-transcribed RNA from the indicated wild-type replicons and selected for 3 weeks with G418 (500 µg ml−1). The resulting cell colonies were stained with crystal violet. S52 GNN with a catalytically inactive mutation in NS5B polymerase was included as a negative control.
Extended Data Figure 3 Effect of SEC14L2 expression on the replication of wild-type and cell culture-adapted HCV subgenomic replicons.
a, b, SEC14L2 expression enhances replication of wild-type HCV RNA in a dose-dependent manner. a, Huh-7.5 cells were transduced with lentiviruses encoding SEC14L2–EGFP fusion protein under a doxycycline-inducible promoter and flow cytometry was performed to obtain single-cell clones. Two cell clones selected for downstream analysis were treated with the indicated concentrations of doxycycline for 24 h, followed by flow cytometry to determine the number of EGFP-positive cells. Mean fluorescence intensities of EGFP are shown at the top of each box. b, S52/SG-neo colony formation in cells described in a. The results were confirmed by two independent transfections. c, SEC14L2 expression enhances replication of cell culture-adapted HCV replicons to varying extents. Colony formation efficiency of the indicated subgenomic replicons in empty-vector- and SEC14L2-expressing Huh-7.5 cells is plotted as CFU per 100,000 transfected cells. Results represent mean ± s.e.m. from two independent transfections.
Extended Data Figure 4 SEC14L2 expression enables replication of wild-type full-length HCV genomes and production of low levels of infectious virus particles.
a, HDFR reporter cells transduced with SEC14L2 or empty vector were transfected with in vitro transcribed RNA from H77, Con1, and J6 full-length genomes. Live cell images were captured 6 days after transfection. White arrows indicate the cells with nuclear RFP. b, The numbers of cells exhibiting nuclear RFP were counted in 3 random microscopic fields at day 6 post-transfection. H77 pol−, with a catalytically inactive mutation in NS5B polymerase, was used as a negative control. c–e, Infectious virus particles are produced from SEC14L2/Huh-7.5 cells harbouring selectable full-length HCV genomes. c, Huh-7.5 cells stably expressing SEC14L2 were incubated at 37 °C for 30 min with anti-CD81 antibody or control IgG, and inoculated with the culture medium from SEC14L2/Huh-7.5 cells harbouring blasticidin-selectable, full-length (FL-BSD) H77, Con1, and J6 genomes, described in Fig. 3b. After 72 h, selection with blasticidin (2.5 µg ml−1) was imposed and the colonies obtained after 3 weeks were stained with crystal violet. d, The cell colonies obtained in c were pooled and stained with anti-NS5A antibody (9E10 clone). The percentages of positive cells from two independent experiments are plotted. As previously described44, 9E10 antibody did not detect NS5A from the J6 isolate. e, The cell colonies obtained in c were pooled and HCV RNA levels were measured by qPCR. The values are plotted as mean ± s.e.m. of two independent experiments.
Extended Data Figure 5 Only full-length SEC14L2 supports replication of wild-type HCV.
a, Huh-7.5 cells stably expressing human or murine SEC14L2 (93% amino acid identity) were lysed and protein expression was confirmed by immunoblotting. These cells were then transfected with S52/SG-neo and selected with G418. The resulting cell colonies were stained with crystal violet. b, c, Only isoform 1 of SEC14L2 supports HCV RNA replication. The SEC14L2 gene is comprised of 12 exons and results in 3 alternatively spliced transcript variants encoding 3 different protein isoforms. Isoform 1 was identified in cDNA screening. b, Schematic representation of SEC14L2 isoforms. Coding exons are shown as green blocks and the amino acid length of each protein is shown on right. c, Cell lysates from Huh-7.5 cells stably expressing 3 SEC14L2 isoforms were analysed by 4–12% SDS–PAGE and immunoblotting was performed with SEC14L2 mouse monoclonal antibody. The bands highlighted with asterisks might be a cleavage product of SEC14L2. UC, untransduced cells. These cells were transfected with S52/SG-neo and selected with G418 for 3 weeks. The resulting cell colonies were stained with crystal violet. d, SEC14L3 and SEC14L4 (86% and 80% amino acid similarity to SEC14L2, respectively)11 are not expressed in Huh-7.5 cells. Cell lysates from Huh-7.5 cells stably expressing SEC14L2, SEC14L3, or SEC14L4 were analysed by 4–12% SDS–PAGE followed by immunoblotting with the indicated antibodies (the signal generated by SEC14L3 antibody in SEC14L2-expressing cells most likely reflects the cross-reactivity of SEC14L3 antibody). These cells were then tested for their ability to support replication of S52/SG-neo. e, f, Deletion mutants of SEC14L2 do not support HCV RNA replication. e, Schematic representation of N-terminal EGFP-tagged SEC14L2 deletion mutants. f, Cell lysates from Huh-7.5 cells stably expressing the full-length SEC14L2 and the deletion mutants were analysed by 4–12% SDS–PAGE followed by immunoblotting with the indicated antibodies. These cells were then transfected with S52/SG-neo and selected with G418 for 3 weeks. The resulting cell colonies were stained with crystal violet. g, h, As N-terminal deletion mutants of SEC14L2 were unstable in Huh-7.5 cells (they formed protein aggregates), we generated chimaeric constructs by fusing C-terminal ends of SEC14L2 with the corresponding N-terminal sequences from SEC14L4 and tested their ability to support HCV replication. g, Schematic representation of C-terminal EGFP-tagged chimaeric constructs. h, Huh-7.5 cells stably expressing the indicated chimaeric constructs were lysed and protein expression was confirmed by immunoblotting with anti-GFP antibody. The cells were then transfected with S52/SG-neo and selected with G418 for 3 weeks.
Extended Data Figure 6 SEC14L2 does not interact with HCV nonstructural proteins under the tested conditions.
a, Yeast two-hybrid assay was performed to examine direct interaction between SEC14L2 and HCV nonstructural proteins. SEC14L2-AD (GAL4 activation domain) fusion or control AD vector was co-expressed with DBD (DNA binding domain) fusion of the individual HCV non-structural proteins or control DBD vector and tested for positive yeast two-hybrid interactions under selective nutritional conditions (lacking leucine, tryptophan, histidine, and adenosine). The strong signal obtained for NS5A most likely reflects the intrinsic trans-activating activity of NS5A45. b, Co-immunoprecipitation did not reveal binding of SEC14L2 with HCVNS5Aprotein. As two-hybrid assay (a) did not yield unambiguous results on interaction between SEC14L2 and NS5A, we employed co-immunoprecipitation assay to probe binding between these proteins. Huh-7.5 cells stably expressing SEC14L2-EGFP and harbouring wild-type Con1/SGneo replicon were lysed and subjected to immunoprecipitation with anti-NS5A antibody, anti-EGFP antibody or control immunoglobulins (IgG). The bound proteins were analysed by immunoblotting. Endogenous MOBKL1B, a previously described binding partner of the NS5A protein41, served as a positive control. c, Subcellular fractionation of SEC14L2-EGFP/Huh-7.5 cells harbouring wild-type Con1/SG-neo replicon did not reveal obvious co-fractionation of SEC14L2 and HCV NS5A protein
Extended Data Figure 7 SEC14L2 does not facilitate HCV RNA replication by modulating PI3K/Akt or cholesterol synthesis pathways.
a–d, Downregulation of PI3K/Akt pathway does not support HCV replication. a, Akt phosphorylation in control and SEC14L2-expressing Huh-7.5 cells was analysed by immunoblotting; β-actin is included as a loading control. b, Stable knockdown of Akt in Huh-7.5 cells with two different shRNAs did not facilitate G418 resistant colony formation by S52/SG-neo. c, Huh-7.5 cells were transduced to stably express PTEN, a negative regulator of PI3K pathway. Despite decreased Akt phosphorylation, these cells did not support colony formation by S52/SG-neo. d, Suppression of PI3K pathway in Huh-7.5 cells by stable expression of a dominant negative Akt, a dominant negative p85 subunit of PI3K, or a constitutively active FOXO3a did not render them permissive to HCV replication. Dominant negative Akt gets phosphorylated, but as it is kinase-dead, it cannot initiate the downstream signalling. Interestingly, increased Akt phosphorylation was seen in Huh-7.5 cells expressing constitutively active FOXO3a, suggesting a potential feedback mechanism. e, f, A SEC14L2 mutant (S289A) lacking the cholesterolgenic activity supported HCV replication. e, Schematic representation of carboxy-terminal EGFP-tagged SEC14L2 point mutants. S288A was generated as a negative control. f, The colony formation efficiency of S52/SG-neo (plotted as CFU per 100,000 transfected cells) and the expression levels of SEC14L2 mutants in Huh-7.5 are shown.
Extended Data Figure 8 SEC14L2 expression masks the effects of lipophilic oxidants and anti-oxidants on H77S.3/GLuc replication.
a, b, SEC14L2 enhances transient replication of H77S.3, but not that of JFH-1 or J6/JFH1. Empty-vector- and SEC14L2-expressing Huh-7.5 cells were electroporated with the indicated viral RNAs lacking GLuc insertions. a, Intracellular and b, extracellular RNA levels were measured 6 days after electroporation. Results are plotted as fold change from empty vector control. c–f, The pro-viral effects of various lipophilic antioxidants on H77S.3/GLuc replication were suppressed in the presence of SEC14L2. c, γ-tocopherol, d, α-tocopheryl succinate, e, α-tocopheryl quinone, and f, sphingosine kinase inhibitor (SKI) were added to Huh-7.5 cells 20 h before transfection with H77S.3/GLuc. Transfections were carried out in the fresh medium lacking these compounds. 6 h after transfection, cells were again fed with each compound and the GLuc expression was measured at 72 h post-transfection. The results are presented as fold change from untreated cells. g–l, SEC14L2 expression suppressed the inhibitory effect of lipophilic oxidants and direct-acting antivirals, but not that of the HCV host factor inhibitors, on H77S.3/GLuc replication. g, Docosahexaenoic acid (oxidant); h, linoleic acid (oxidant); i, interferon-β (IFN-β); j, CSA (cyclosporine A); k, danoprevir (NS3 protease inhibitor) and l, 2′CMeA (NS5B polymerase inhibitor) were added to Huh-7.5 cells 6 h after transfection with H77S.3/GLuc (and Jc1/GLuc in the case of Danoprevir and 2′CMeA) and the secreted GLuc activity at 72 h post-transfection was measured. The results are plotted as percentage of inhibition relative to the untreated cells. All results in this figure represent mean ± s.d. of two replicate experiments.
Extended Data Figure 9 SEC14L2 expression does not enhance replication of lipid peroxidation-resistant RNA viruses.
a–f, Flaviviruses, such as yellow fever virus (YFV) and dengue virus (DENV) do not respond to SEC14L2 expression. a–e, Empty-vector- and SEC14L2-expressing Huh-7.5 cells were infected with YFV-venus (a, b) or DENV-GFP (d, e) at a multiplicity of infection (MOI) of 0.01 and 0.1, respectively. The cells were harvested at the indicated times post-infection, followed by FACS analysis. a, d, The number of yellow (YFV-venus) and green (DENV-GFP) cells are plotted as percentage of positive cells and b, e, the mean fluorescence intensities (MFI) are presented as percentage relative to empty vector. c, f, Empty-vector- and SEC14L2-expressing Huh-7.5 cells were inoculated with c, YFV (17D) or f, DENV (serotype 2 strain 16681) at an MOI of 0.5 and 0.1, respectively. The culture medium was collected at the indicated times post infection and the infectious virus titres were determined by a plaque formation assay on Huh-7.5 cells (YFV-venus) or BHK cells (DENV). The results are presented as plaque-forming units (PFU) per ml. g–j, Alphaviruses, such as Sindbis virus (SINV) and Ross River virus (RRV) are insensitive to SEC14L2 expression. Empty-vector- and SEC14L2-expressing Huh-7.5 cells were infected with SINV-GFP (g, h) or RRV-GFP (i, j) at an MOI of 0.05. The cells were harvested at the indicated times post-infection and FACS analysis was carried out to determine the number of green cells and the mean fluorescence intensities. All results in this figure represent mean ± s.e.m. of two replicate experiments.
Extended Data Figure 10 VE increased SEC14L2-mediated replicon colony formation.
a, VE (α-tocopherol, 1 µM) was added to Huh-7.5 cells 20 h before transfection with the indicated wild-type HCV subgenomic replicons. Transfections were carried out for 6 h in the fresh medium lacking VE, followed by medium change to VE-containing medium. After 48 h, cells were subjected to G418 selection and fed with fresh G418 and VE every 2 days. The resulting cell colonies were stained with crystal violet. Shown are the results of one of three independent experiments. b, Colony formation efficiency of the indicated subgenomic replicons was measured in SEC14L2-expressing cells in the absence or presence of 1 µM VE. The results are plotted as mean ± s.e.m. of CFU per 100,000 transfected cells from three independent transfections. **P < 0.005 by two-tailed, paired t-test.
Rights and permissions
About this article
Cite this article
Saeed, M., Andreo, U., Chung, HY. et al. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature 524, 471–475 (2015). https://doi.org/10.1038/nature14899
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14899
This article is cited by
-
Identification of SEC14 like lipid binding 2(SEC14L2) sequence and expression profiles in the Chinese tree shrew (Tupaia belangeri chinensis)
Molecular Biology Reports (2022)
-
A Golgi-derived vesicle potentiates PtdIns4P to PtdIns3P conversion for endosome fission
Nature Cell Biology (2021)
-
Hepatitis C virus cell culture models: an encomium on basic research paving the road to therapy development
Medical Microbiology and Immunology (2019)
-
Expression of human CD46 and trans-complementation by murine adenovirus 1 fails to allow productive infection by a group B oncolytic adenovirus in murine cancer cells
Journal for ImmunoTherapy of Cancer (2018)
-
NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR
Nature Immunology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.