Abstract
Bacteria secrete peptides and proteins to communicate, to poison competitors, and to manipulate host cells. Among the various protein-translocation machineries, the peptidase-containing ATP-binding cassette transporters (PCATs) are appealingly simple. Each PCAT contains two peptidase domains that cleave the secretion signal from the substrate, two transmembrane domains that form a translocation pathway, and two nucleotide-binding domains that hydrolyse ATP. In Gram-positive bacteria, PCATs function both as maturation proteases and exporters for quorum-sensing or antimicrobial polypeptides. In Gram-negative bacteria, PCATs interact with two other membrane proteins to form the type 1 secretion system. Here we present crystal structures of PCAT1 from Clostridium thermocellum in two different conformations. These structures, accompanied by biochemical data, show that the translocation pathway is a large α-helical barrel sufficient to accommodate small folded proteins. ATP binding alternates access to the transmembrane pathway and also regulates the protease activity, thereby coupling substrate processing to translocation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Håvarstein, L. S., Diep, D. B. & Nes, I. F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16, 229–240 (1995)
ter Beek, J., Guskov, A. & Slotboom, D. J. Structural diversity of ABC transporters. J. Gen. Physiol. 143, 419–435 (2014)
Rice, A. J., Park, A. & Pinkett, H. W. Diversity in ABC transporters: type I, II and III importers. Crit. Rev. Biochem. Mol. Biol. 49, 426–437 (2014)
Rawlings, N. D., Waller, M., Barrett, A. J. & Bateman, A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 (2014)
Gebhard, S. ABC transporters of antimicrobial peptides in Firmicutes bacteria - phylogeny, function and regulation. Mol. Microbiol. 86, 1295–1317 (2012)
Lenders, M. H., Reimann, S., Smits, S. H. & Schmitt, L. Molecular insights into type I secretion systems. Biol. Chem. 394, 1371–1385 (2013)
Lecher, J. et al. An RTX transporter tethers its unfolded substrate during secretion via a unique N-terminal domain. Structure 20, 1778–1787 (2012)
Ishii, S. et al. Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway. J. Biol. Chem. 285, 10777–10785 (2010)
Ishii, S., Yano, T., Okamoto, A., Murakawa, T. & Hayashi, H. Boundary of the nucleotide-binding domain of Streptococcus ComA based on functional and structural analysis. Biochemistry 52, 2545–2555 (2013)
Zaitseva, J. et al. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 25, 3432–3443 (2006)
Schmitt, L., Benabdelhak, H., Blight, M. A., Holland, I. B. & Stubbs, M. T. Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: identification of a variable region within ABC helical domains. J. Mol. Biol. 330, 333–342 (2003)
Zaitseva, J., Jenewein, S., Jumpertz, T., Holland, I. B. & Schmitt, L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 24, 1901–1910 (2005)
Gentschev, I. & Goebel, W. Topological and functional studies on HlyB of Escherichia coli. Mol. Gen. Genet. 232, 40–48 (1992)
Koronakis, V. & Hughes, C. Bacterial signal peptide-independent protein export: HlyB-directed secretion of hemolysin. Semin. Cell Biol. 4, 7–15 (1993)
Orelle, C., Dalmas, O., Gros, P., Di Pietro, A. & Jault, J. M. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 278, 47002–47008 (2003)
Moody, J. E., Millen, L., Binns, D., Hunt, J. F. & Thomas, P. J. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 277, 21111–21114 (2002)
Sissons, C. H., Sharrock, K. R., Daniel, R. M. & Morgan, H. W. Isolation of cellulolytic anaerobic extreme thermophiles from New Zealand thermal sites. Appl. Environ. Microbiol. 53, 832–838 (1987)
Gorbulev, S., Abele, R. & Tampe, R. Allosteric crosstalk between peptide-binding, transport, and ATP hydrolysis of the ABC transporter TAP. Proc. Natl Acad. Sci. USA 98, 3732–3737 (2001)
Davidson, A. L., Shuman, H. A. & Nikaido, H. Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc. Natl Acad. Sci. USA 89, 2360–2364 (1992)
Rosenberg, M. F., Kamis, A. B., Aleksandrov, L. A., Ford, R. C. & Riordan, J. R. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR). J. Biol. Chem. 279, 39051–39057 (2004)
Li, C. et al. ATPase activity of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 271, 28463–28468 (1996)
Ujwal, R. & Abramson, J. High-throughput crystallization of membrane proteins using the lipidic bicelle method. J. Vis. Exp. 59, e3383 (2012)
Oldham, M. L. & Chen, J. Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 332, 1202–1205 (2011)
Khare, D., Oldham, M. L., Orelle, C., Davidson, A. L. & Chen, J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol. Cell 33, 528–536 (2009)
Wu, K. H. & Tai, P. C. Cys32 and His105 are the critical residues for the calcium-dependent cysteine proteolytic activity of CvaB, an ATP-binding cassette transporter. J. Biol. Chem. 279, 901–909 (2004)
Ishii, S., Yano, T. & Hayashi, H. Expression and characterization of the peptidase domain of Streptococcus pneumoniae ComA, a bifunctional ATP-binding cassette transporter involved in quorum sensing pathway. J. Biol. Chem. 281, 4726–4731 (2006)
van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004)
Park, E. et al. Structure of the SecY channel during initiation of protein translocation. Nature 506, 102–106 (2014)
Gogala, M. et al. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107–110 (2014)
Zimmer, J., Nam, Y. & Rapoport, T. A. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 (2008)
Egea, P. F. & Stroud, R. M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl Acad. Sci. USA 107, 17182–17187 (2010)
Mingarro, I., Nilsson, I., Whitley, P. & von Heijne, G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 1, 3 (2000)
Kowarik, M., Kung, S., Martoglio, B. & Helenius, A. Protein folding during cotranslational translocation in the endoplasmic reticulum. Mol. Cell 10, 769–778 (2002)
Faham, S. & Bowie, J. U. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 316, 1–6 (2002)
Chen, S., Oldham, M. L., Davidson, A. L. & Chen, J. Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature 499, 364–368 (2013)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
French, G. S. & Wilson, K. S. On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 (1978)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Afonine, P. V., Grosse-Kunstleve, R. W. & Adams, P. D. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. D 61, 850–855 (2005)
Painter, J. & Merritt, E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 (2006)
Schröder, G. F., Levitt, M. & Brunger, A. T. Super-resolution biomolecular crystallography with low-resolution data. Nature 464, 1218–1222 (2010)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
DeLano, W. L. in The PyMOL User’s Manual (DeLano Scientific, 2002)
Heginbotham, L., LeMasurier, M., Kolmakova-Partensky, L. & Miller, C. Single Streptomyces lividans K+ channels: functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114, 551–560 (1999)
Long, S. B., Tao, X., Campbell, E. B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007)
Tao, X. & MacKinnon, R. Functional analysis of Kv1.2 and paddle chimera Kv channels in planar lipid bilayers. J. Mol. Biol. 382, 24–33 (2008)
Brohawn, S. G., del Marmol, J. & MacKinnon, R. Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science 335, 436–441 (2012)
Scharschmidt, B. F., Keeffe, E. B., Blankenship, N. M. & Ockner, R. K. Validation of a recording spectrophotometric method for measurement of membrane-associated Mg- and NaK-ATPase activity. J. Lab. Clin. Med. 93, 790–799 (1979)
Orelle, C., Ayvaz, T., Everly, R. M., Klug, C. S. & Davidson, A. L. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc. Natl Acad. Sci. USA 105, 12837–12842 (2008)
Acknowledgements
We thank the staff at the Advance Photon Source GM/CA-CAT and NE-CAT for assistance with data collection, S. McCarry for editing the manuscript, R. MacKinnon and D. Kearns for helpful discussions, M. L. Oldham for help with figure preparation, and H. Zhang and W. Mi for efforts in the early stage of this project. This work was supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
All authors designed the study and analysed the data. D.Y.-w.L. and S. H. performed cloning and biochemical experiments. D.Y.-w.L. determined the crystal structures and wrote the manuscript together with J.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
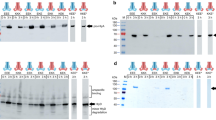
Extended Data Figure 2 PCAT1 protease activities towards substrates of other Gram-positive bacteria.
PCAT1 was able to cleave its putative substrate, Cthe_0535, from C. thermocellum at 37 °C for 2 h but showed no proteolytic activities towards CA_P0072 from Clostridium acetobutylicum or ComC from Streptococcus pneumoniae.
Extended Data Figure 3 Anomalous difference Fourier electron density map.
Stereoview of the backbone of SeMet-substituted PCAT1 (grey ribbon). Methionine residues are shown in orange sticks. The blue mesh contoured at 3σ represents the superimposed anomalous difference Fourier map calculated from data collected on four different PCAT1 constructs. A total of 28 selenium sites were identified and used as markers to assist assignment of the sequence register. Out of the 21 native methionine residues, only two were not identified (Met 1 and Met 271), probably reflecting the conformational flexibility of these residues.
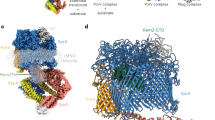
Extended Data Figure 5 The TM tunnel in the ATP-free form is large enough to accommodate a small protein.
The bovine pancreatic trypsin inhibitor (PDB accession 4PTI) is modelled into the TM tunnel of PCAT1, shown as a blue ribbon, to illustrate the size of the cavity.
Rights and permissions
About this article
Cite this article
Lin, Dw., Huang, S. & Chen, J. Crystal structures of a polypeptide processing and secretion transporter. Nature 523, 425–430 (2015). https://doi.org/10.1038/nature14623
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14623
This article is cited by
-
Structural basis of peptide secretion for Quorum sensing by ComA
Nature Communications (2023)
-
A step forward to the optimized HlyA type 1 secretion system through directed evolution
Applied Microbiology and Biotechnology (2023)
-
Mechanisms of membrane protein crystallization in ‘bicelles’
Scientific Reports (2022)
-
Recent Insights into the Structure and Function of Mycobacterial Membrane Proteins Facilitated by Cryo-EM
The Journal of Membrane Biology (2021)
-
Impact of the nisin modification machinery on the transport kinetics of NisT
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.