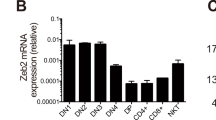
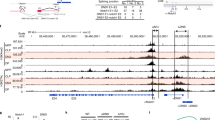
Abstract
The vertebrate thymus provides an inductive environment for T-cell development. Within the mouse thymus, Notch signals are indispensable for imposing the T-cell fate on multipotential haematopoietic progenitors, but the downstream effectors that impart T-lineage specification and commitment are not well understood. Here we show that a transcription factor, T-cell factor 1 (TCF-1; also known as transcription factor 7, T-cell specific, TCF7), is a critical regulator in T-cell specification. TCF-1 is highly expressed in the earliest thymic progenitors, and its expression is upregulated by Notch signals. Most importantly, when TCF-1 is forcibly expressed in bone marrow (BM) progenitors, it drives the development of T-lineage cells in the absence of T-inductive Notch1 signals. Further characterization of these TCF-1-induced cells revealed expression of many T-lineage genes, including T-cell-specific transcription factors Gata3 and Bcl11b, and components of the T-cell receptor. Our data suggest a model where Notch signals induce TCF-1, and TCF-1 in turn imprints the T-cell fate by upregulating expression of T-cell essential genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schwarz, B. A. et al. Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 178, 2008–2017 (2007)
Spangrude, G. J. & Scollay, R. Differentiation of hematopoietic stem cells in irradiated mouse thymic lobes. Kinetics and phenotype of progeny. J. Immunol. 145, 3661–3668 (1990)
Doulatov, S. et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nature Immunol. 11, 585–593 (2010)
Rothenberg, E. V., Zhang, J. & Li, L. Multilayered specification of the T-cell lineage fate. Immunol. Rev. 238, 150–168 (2010)
Pui, J. C. et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299–308 (1999)
Radtke, F. et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1 . Immunity 10, 547–558 (1999)
Sambandam, A. et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nature Immunol. 6, 663–670 (2005)
Taghon, T. N., David, E. S., Zuniga-Pflucker, J. C. & Rothenberg, E. V. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 19, 965–978 (2005)
Verbeek, S. et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374, 70–74 (1995)
Schilham, M. W. et al. Critical involvement of Tcf-1 in expansion of thymocytes. J. Immunol. 161, 3984–3991 (1998)
Goux, D. et al. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood 106, 1726–1733 (2005)
Okamura, R. M. et al. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity 8, 11–20 (1998)
Schmitt, T. M. & Zuniga-Pflucker, J. C. T-cell development, doing it in a dish. Immunol. Rev. 209, 95–102 (2006)
Huang, J. et al. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with Notch receptor ligation. J. Immunol. 175, 4858–4865 (2005)
Cobas, M. et al. β-catenin is dispensable for hematopoiesis and lymphopoiesis. J. Exp. Med. 199, 221–229 (2004)
Jeannet, G. et al. Long-term, multilineage hematopoiesis occurs in the combined absence of β-catenin and γ-catenin. Blood 111, 142–149 (2008)
Tago, K. et al. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev. 14, 1741–1749 (2000)
Pear, W. S. et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183, 2283–2291 (1996)
Deftos, M. L. et al. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13, 73–84 (2000)
van de Wetering, M., Oosterwegel, M., Dooijes, D. & Clevers, H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10, 123–132 (1991)
Yu, Q. et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-γ. Nature Immunol. 10, 992–999 (2009)
Hosoya, T. et al. GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 206, 2987–3000 (2009)
Ikawa, T. et al. An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 (2010)
Li, L., Leid, M. & Rothenberg, E. V. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b . Science 329, 89–93 (2010)
Li, P. et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85–89 (2010)
Taghon, T. et al. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity 24, 53–64 (2006)
Yashiro-Ohtani, Y. et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 23, 1665–1676 (2009)
Lin, Y. C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nature Immunol. 11, 635–643 (2010)
Wendorff, A. A. et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 33, 671–684 (2010)
Liu, Z. et al. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J. Clin. Invest. 121, 800–808 (2011)
Brault, V. et al. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264 (2001)
Huang, J. et al. Pivotal role for glycogen synthase kinase-3 in hematopoietic stem cell homeostasis in mice. J. Clin. Invest. 119, 3519–3529 (2009)
Knudsen, S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics 15, 356–361 (1999)
Weng, A. P. et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109 (2006)
Pavlidis, P. & Noble, W. S. Matrix2png: a utility for visualizing matrix data. Bioinformatics 19, 295–296 (2003)
Acknowledgements
We thank H. Clevers for permission to use TCF-1−/− mice and W. Pear, S. Reiner, C. Bassing, R. Sen and W. Bailis for critical comments. This work was supported by NIH grant AI059621 and a Scholar Award from the Leukemia and Lymphoma Society (A.B.), and institutional training grants T32AI055428 (B.N.W.) and T32CA09140 (A.W.-S.C.).
Author information
Authors and Affiliations
Contributions
B.N.W., A.W.-S.C. and A.B. designed and performed the experiments. Q.Y., Y.Y.-O. and A.C. helped with experiments. O.S. provided mice and technical assistance. B.N.W. and A.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-13 with legends, additional references and Supplementary Tables 1-3. (PDF 4250 kb)
Rights and permissions
About this article
Cite this article
Weber, B., Chi, AS., Chavez, A. et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011). https://doi.org/10.1038/nature10279
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10279
This article is cited by
-
Canalizing cell fate by transcriptional repression
Molecular Systems Biology (2024)
-
Intrinsically disordered domain of transcription factor TCF-1 is required for T cell developmental fidelity
Nature Immunology (2023)
-
Intrathymic dendritic cell-biased precursors promote human T cell lineage specification through IRF8-driven transmembrane TNF
Nature Immunology (2023)
-
TCF-1 regulates NKG2D expression on CD8 T cells during anti-tumor responses
Cancer Immunology, Immunotherapy (2023)
-
Aiolos represses CD4+ T cell cytotoxic programming via reciprocal regulation of TFH transcription factors and IL-2 sensitivity
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.