Abstract
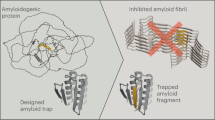
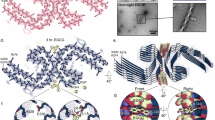
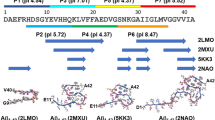
Many globular and natively disordered proteins can convert into amyloid fibrils. These fibrils are associated with numerous pathologies1 as well as with normal cellular functions2,3, and frequently form during protein denaturation4,5. Inhibitors of pathological amyloid fibril formation could be useful in the development of therapeutics, provided that the inhibitors were specific enough to avoid interfering with normal processes. Here we show that computer-aided, structure-based design can yield highly specific peptide inhibitors of amyloid formation. Using known atomic structures of segments of amyloid fibrils as templates, we have designed and characterized an all-d-amino-acid inhibitor of the fibril formation of the tau protein associated with Alzheimer’s disease, and a non-natural l-amino-acid inhibitor of an amyloid fibril that enhances sexual transmission of human immunodeficiency virus. Our results indicate that peptides from structure-based designs can disrupt the fibril formation of full-length proteins, including those, such as tau protein, that lack fully ordered native structures. Because the inhibiting peptides have been designed on structures of dual-β-sheet ‘steric zippers’, the successful inhibition of amyloid fibril formation strengthens the hypothesis that amyloid spines contain steric zippers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Westermark, P. et al. A primer of amyloid nomenclature. Amyloid 14, 179–183 (2007)
Maji, S. K. et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 (2009)
Fowler, D. M., Koulov, A. V., Balch, W. E. & Kelly, J. W. Functional amyloid – from bacteria to humans. Trends Biochem. Sci. 32, 217–224 (2007)
Astbury, W. T. & Dickinson, S. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 29, 2351–2360 (1935)
Calamai, M., Chiti, F. & Dobson, C. M. Amyloid fibril formation can proceed from different conformations of a partially unfolded protein. Biophys. J. 89, 4201–4210 (2005)
Tjernberg, L. O. et al. Arrest of β-amyloid fibril formation by a pentapeptide ligand. J. Biol. Chem. 271, 8545–8548 (1996)
Findeis, M. A. Peptide inhibitors of β amyloid aggregation. Curr. Top. Med. Chem. 2, 417–423 (2002)
Sciarretta, K. L., Gordon, D. J. & Meredith, S. C. Peptide-based inhibitors of amyloid assembly. Methods Enzymol. 413, 273–312 (2006)
Soto, C., Kindy, M. S., Baumann, M. & Frangione, B. Inhibition of Alzheimer’s amyloidosis by peptides that prevent β-sheet conformation. Biochem. Biophys. Res. Commun. 226, 672–680 (1996)
Kokkoni, N., Stott, K., Amijee, H., Mason, J. M. & Doig, A. J. N-methylated peptide inhibitors of amyloid aggregation and toxicity. Optimization of the inhibitor structure. Biochemistry 45, 9906–9918 (2006)
Sato, T. et al. Inhibitors of amyloid toxicity based on β-sheet packing of Aβ40 and Aβ42. Biochemistry 45, 5503–5516 (2006)
Larbig, G., Pickhardt, M., Lloyd, D. G., Schmidt, B. & Mandelkow, E. Screening for inhibitors of tau protein aggregation into Alzheimer paired helical filaments: a ligand based approach results in successful scaffold hopping. Curr. Alzheimer Res. 4, 315–323 (2007)
Wiesehan, K. et al. Selection of D-amino-acid peptides that bind to Alzheimer’s disease amyloid peptide aβ1–42 by mirror image phage display. ChemBioChem 4, 748–753 (2003)
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005)
Sawaya, M. R. et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447, 453–457 (2007)
Wiltzius, J. J. et al. Molecular mechanisms for protein-encoded inheritance. Nature Struct. Mol. Biol. 16, 973–978 (2009)
Selkoe, D. J. Alzheimer’s disease: genes, proteins, and therapy. Physiol. Rev. 81, 741–766 (2001)
Goux, W. J. et al. The formation of straight and twisted filaments from short tau peptides. J. Biol. Chem. 279, 26868–26875 (2004)
von Bergen, M. et al. Assembly of τ protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc. Natl Acad. Sci. USA 97, 5129–5134 (2000)
Goldschmidt, L., Teng, P. K., Riek, R. & Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl Acad. Sci. USA 107, 3487–3492 (2010)
Thompson, M. J. et al. The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl Acad. Sci. USA 103, 4074–4078 (2006)
Münch, J. et al. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 131, 1059–1071 (2007)
Friedhoff, P., von Bergen, M., Mandelkow, E. M., Davies, P. & Mandelkow, E. A nucleated assembly mechanism of Alzheimer paired helical filaments. Proc. Natl Acad. Sci. USA 95, 15712–15717 (1998)
Wille, H., Drewes, G., Biernat, J., Mandelkow, E. M. & Mandelkow, E. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro . J. Cell Biol. 118, 573–584 (1992)
Kuhlman, B. et al. Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 (2003)
Chen, Z., Krause, G. & Reif, B. Structure and orientation of peptide inhibitors bound to β-amyloid fibrils. J. Mol. Biol. 354, 760–776 (2005)
Roan, N. R. et al. The cationic properties of SEVI underlie its ability to enhance human immunodeficiency virus infection. J. Virol. 83, 73–80 (2009)
Petrassi, H. M., Klabunde, T., Sacchettini, J. & Kelly, J. W. Structure-based design of N-phenyl phenoxazine transthyretin amyloid fibril inhibitors. J. Am. Chem. Soc. 122, 2178–2192 (2000)
Schweers, O., Schonbrunn-Hanebeck, E., Marx, A. & Mandelkow, E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for β-structure. J. Biol. Chem. 269, 24290–24297 (1994)
Fleishman, S. J. et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 332, 816–821 (2011)
Collaborative Computational Project, Number 4 . The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Lawrence, M. C. & Colman, P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 (1993)
Studier, F. W., Rosenberg, A. H., Dunn, J. J. & Dubendorff, J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60–89 (1990)
Biernat, J. et al. The switch of tau protein to an Alzheimer-like state includes the phosphorylation of two serine-proline motifs upstream of the microtubule binding region. EMBO J. 11, 1593–1597 (1992)
Barghorn, S., Biernat, J. & Mandelkow, E. Purification of recombinant tau protein and preparation of Alzheimer-paired helical filaments in vitro. Methods Mol. Biol. 299, 35–51 (2005)
Friedhoff, P., Schneider, A., Mandelkow, E. M. & Mandelkow, E. Rapid assembly of Alzheimer-like paired helical filaments from microtubule-associated protein tau monitored by fluorescence in solution. Biochemistry 37, 10223–10230 (1998)
Pérez, M., Valpuesta, J. M., Medina, M., Montejo de Garcini, E. & Avila, J. Polymerization of tau into filaments in the presence of heparin: the minimal sequence required for tau-tau interaction. J. Neurochem. 67, 1183–1190 (1996)
Schweers, O., Mandelkow, E. M., Biernat, J. & Mandelkow, E. Oxidation of cysteine-322 in the repeat domain of microtubule-associated protein tau controls the in vitro assembly of paired helical filaments. Proc. Natl Acad. Sci. USA 92, 8463–8467 (1995)
R Development Core Team . R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna) 〈http://www.r-project.org〉 (2008)
Rojas Quijano, F. A., Morrow, D., Wise, B. M., Brancia, F. L. & Goux, W. J. Prediction of nucleating sequences from amyloidogenic propensities of tau-related peptides. Biochemistry 45, 4638–4652 (2006)
Morris, A. M., Watzky, M. A., Agar, J. N. & Finke, R. G. Fitting neurological protein aggregation kinetic data via a 2-step, minimal “Ockham’s razor” model: the Finke-Watzky mechanism of nucleation followed by autocatalytic surface growth. Biochemistry 47, 2413–2427 (2008)
Schmidt, K., Segond von Banchet, G. & Heppelmann, B. Labelling of peptides with 1.4-nm gold particles to demonstrate their binding sites in the rat spinal cord. J. Neurosci. Methods 87, 195–200 (1999)
Eisenberg, D., Wesson, M. & Yamashita, M. Interpretation of protein folding and binding with atomic solvation parameters. Chem. Scr. 29A, 217–221 (1989)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
DeLano, W. L. PyMOL Molecular Viewer 〈http://www.pymol.org〉 (2002)
Papkalla, A., Munch, J., Otto, C. & Kirchhoff, F. Nef enhances human immunodeficiency virus type 1 infectivity and replication independently of viral coreceptor tropism. J. Virol. 76, 8455–8459 (2002)
Platt, E. J., Wehrly, K., Kuhmann, S. E., Chesebro, B. & Kabat, D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72, 2855–2864 (1998)
Acknowledgements
We thank M. I. Ivanova, J. Corn, T. Kortemme, D. Anderson, M. R. Sawaya, M. Phillips, S. Sambashivan, J. Park, M. Landau, A. Laganowsky, Q. Zhang, R. Clubb, F. Guo, T. Yeates, J. Nowick, J. Zheng and M. J. Thompson for discussions; the HHMI, NIH, NSF, Gates Foundation and Joint Center for Translational Medicine for support; R. Peterson for help with NMR experiments; E. Mandelkow for providing tau constructs; R. Riek for providing amyloid-β; and J. Stroud for amyloid-β preparation. Support came from the Damon Runyon Cancer Research Foundation (J.K.), the Ruth L. Kirschstein National Research Service Award (H.W.C.), the programme for junior professors by the Ministry of Science, Baden-Württemberg (J.M.), and a UCLA-IGERT bioinformatics traineeship (S.A.S.).
Author information
Authors and Affiliations
Contributions
S.A.S., J.K., D.B., J.M. and D.E. designed the project. J.K. and S.A.S. created the design protocol. J.K. designed the d-peptides. L.J. expanded the design methodology and designed the non-natural amino-acid peptides. S.A.S., H.W.C. and A.Z. performed the fluorescence experiments and electron microscopy, and analysed kinetic data. A.Z. determined the structure of GGVLVN. O.Z. performed the HIV infectivity experiments. J.T.S. determined the tau fibril elongation rates. S.A.S. performed the NMR experiments. S.A.S., J.K. and D.E. wrote the manuscript and coordinated contributions by other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-22 with legends and Supplementary Tables 1-2. (PDF 3543 kb)
Rights and permissions
About this article
Cite this article
Sievers, S., Karanicolas, J., Chang, H. et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475, 96–100 (2011). https://doi.org/10.1038/nature10154
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10154
This article is cited by
-
A novel transgenic mouse line with hippocampus-dominant and inducible expression of truncated human tau
Translational Neurodegeneration (2023)
-
Sequence-based prediction of the intrinsic solubility of peptides containing non-natural amino acids
Nature Communications (2023)
-
Tau Aggregation Inhibiting Peptides as Potential Therapeutics for Alzheimer Disease
Cellular and Molecular Neurobiology (2023)
-
Identification of potential inhibitors against Alzheimer-related proteins in Cordyceps militaris ethanol extract: experimental evidence and computational analyses
3 Biotech (2023)
-
Simulations of Amyloid-Forming Peptides in the Crystal State
The Protein Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.