Abstract
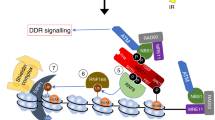
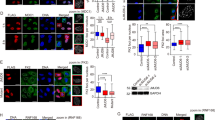
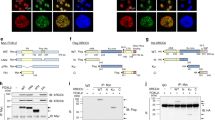
p53-binding protein 1 (53BP1) is known to be an important mediator of the DNA damage response1, with dimethylation of histone H4 lysine 20 (H4K20me2) critical to the recruitment of 53BP1 to double-strand breaks (DSBs)2,3. However, it is not clear how 53BP1 is specifically targeted to the sites of DNA damage, as the overall level of H4K20me2 does not seem to increase following DNA damage. It has been proposed that DNA breaks may cause exposure of methylated H4K20 previously buried within the chromosome; however, experimental evidence for such a model is lacking. Here we found that H4K20 methylation actually increases locally upon the induction of DSBs and that methylation of H4K20 at DSBs is mediated by the histone methyltransferase MMSET (also known as NSD2 or WHSC1) in mammals. Downregulation of MMSET significantly decreases H4K20 methylation at DSBs and the subsequent accumulation of 53BP1. Furthermore, we found that the recruitment of MMSET to DSBs requires the γH2AX–MDC1 pathway; specifically, the interaction between the MDC1 BRCT domain and phosphorylated Ser 102 of MMSET. Thus, we propose that a pathway involving γH2AX–MDC1–MMSET regulates the induction of H4K20 methylation on histones around DSBs, which, in turn, facilitates 53BP1 recruitment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
FitzGerald, J. E., Grenon, M. & Lowndes, N. F. 53BP1: function and mechanisms of focal recruitment. Biochem. Soc. Trans. 37, 897–904 (2009)
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006)
Sanders, S. L. et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 (2004)
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 5, 675–679 (2003)
Bekker-Jensen, S., Lukas, C., Melander, F., Bartek, J. & Lukas, J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170, 201–211 (2005)
Lou, Z. et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187–200 (2006)
Stewart, G. S., Wang, B., Bignell, C. R., Taylor, A. M. & Elledge, S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 (2003)
Du, L. L., Nakamura, T. M. & Russell, P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 20, 1583–1596 (2006)
Moynahan, M. E., Pierce, A. J. & Jasin, M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 (2001)
Fang, J. et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12, 1086–1099 (2002)
Nishioka, K. et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9, 1201–1213 (2002)
Schotta, G. et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004)
Schotta, G. et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22, 2048–2061 (2008)
Kim, J. Y. et al. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol. Cell. Biol. 28, 2023–2034 (2008)
Marango, J. et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 111, 3145–3154 (2008)
Nimura, K. et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 460, 287–291 (2009)
Bergemann, A. D., Cole, F. & Hirschhorn, K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 21, 188–195 (2005)
Chesi, M. et al. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92, 3025–3034 (1998)
Stec, I. et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet. 7, 1071–1082 (1998)
Wang, B., Matsuoka, S., Carpenter, P. B. & Elledge, S. J. 53BP1, a mediator of the DNA damage checkpoint. Science 298, 1435–1438 (2002)
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007)
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007)
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007)
Stucki, M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 (2005)
Yu, X., Chini, C. C., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003)
Manke, I. A., Lowery, D. M., Nguyen, A. & Yaffe, M. B. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302, 636–639 (2003)
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007)
Lee, M. S., Edwards, R. A., Thede, G. L. & Glover, J. N. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the γ-H2AX histone tail. J. Biol. Chem. 280, 32053–32056 (2005)
Kim, J. E., Chen, J. & Lou, Z. DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 (2008)
You, Z. et al. CtIP links DNA double-strand break sensing to resection. Mol. Cell 36, 954–969 (2009)
Acknowledgements
We thank M. Jasin for providing HeLa DR–GFP cells, and S. Baylin for providing MDA-MB-231 cells with inducible I-SceI expression. We thank M. Goldberg, X. Yu and M. Stucki for providing MDC1 and BRCA1 constructs. We thank M. Huen for providing 53BP1 expression constructs. We thank J. D. Licht for providing MMSET antibodies and B. Ho Park for providing MMSET shRNAs. This work was supported by the Richard Schulze Family Foundation and the NIH (CA130996 and CA151329; Z.L.), a grant from the American Cancer Society (IRG-58-010-52; Z.Y.) and a Siteman Career Award in Breast Cancer Research (Z.Y.).
Author information
Authors and Affiliations
Contributions
H.P. designed and performed the experiments; L.Z., K.L., Y.Q. and F.F. performed some experiments; M.C. and P.L.B. provided essential tools; L.W., Z.Y. and Z.L. designed the experiments and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
The file contains Supplementary Figures 1-5 with legends and Supplementary Figures 1-4 for the raw data. (PDF 3329 kb)
Rights and permissions
About this article
Cite this article
Pei, H., Zhang, L., Luo, K. et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 (2011). https://doi.org/10.1038/nature09658
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09658
This article is cited by
-
Loss of p53 function promotes DNA damage-induced formation of nuclear actin filaments
Cell Death & Disease (2023)
-
Roles for the methyltransferase SETD8 in DNA damage repair
Clinical Epigenetics (2022)
-
Dynamic recruitment of UFM1-specific peptidase 2 to the DNA double-strand breaks regulated by WIP1
Genome Instability & Disease (2022)
-
Biological function and regulation of histone 4 lysine 20 methylation in DNA damage response
Genome Instability & Disease (2022)
-
The role of NSD1, NSD2, and NSD3 histone methyltransferases in solid tumors
Cellular and Molecular Life Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.