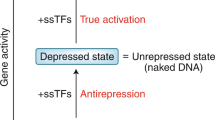
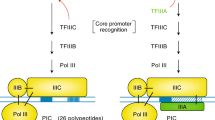
Abstract
In the eukaryotic genome, the thousands of genes that encode messenger RNA are transcribed by a molecular machine called RNA polymerase II. Analysing the distribution and status of RNA polymerase II across a genome has provided crucial insights into the long-standing mysteries of transcription and its regulation. These studies identify points in the transcription cycle where RNA polymerase II accumulates after encountering a rate-limiting step. When coupled with genome-wide mapping of transcription factors, these approaches identify key regulatory steps and factors and, importantly, provide an understanding of the mechanistic generalities, as well as the rich diversities, of gene regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Juven-Gershon, T., Hsu, J. Y., Theisen, J. W. & Kadonaga, J. T. The RNA polymerase II core promoter — the gateway to transcription. Curr. Opin. Cell Biol. 20, 253–259 (2008).
Lee, T. I. et al. Transcriptional regulatory networks in Saccharomyces cerevisiae . Science 298, 799–804 (2002).
Saunders, A., Core, L. J. & Lis, J. T. Breaking barriers to transcription elongation. Nature Rev. Mol. Cell Biol. 7, 557–567 (2006).
Venters, B. J. & Pugh, B. F. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19, 360–371 (2009). This paper examines the genome-wide distribution of Pol II in yeast and suggests that yeast have Pol II enrichment on the 59 ends of many genes.
Muse, G. W. et al. RNA polymerase is poised for activation across the genome. Nature Genet. 39, 1507–1511 (2007). This paper examines the genome-wide distribution of Pol II in Drosophila S2 cells and demonstrates that there is Pol II enrichment on many promoters in a stalled or paused state.
Zeitlinger, J. et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature Genet. 39, 1512–1516 (2007). This paper examines the genome-wide distribution of Pol II in Drosophila embryos and suggests that promoter-proximal stalling/pausing occurs at many developmental genes.
Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R. & Young, R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007). This paper examines the promoter architecture in human cells and suggests that there is Pol II enrichment on promoters and that Pol II seems to have initiated transcription at most genes.
Ptashne, M. & Gann, A. Transcriptional activation by recruitment. Nature 386, 569–577 (1997). This paper reviews the evidence for regulation by Pol II recruitment in bacteria and yeast.
Mirkovitch, J. & Darnell, J. Jr. Mapping of RNA polymerase on mammalian genes in cells and nuclei. Mol. Biol. Cell 3, 1085–1094 (1992).
Rougvie, A. E. & Lis, J. T. The RNA polymerase II molecule at the 59 end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795–804 (1988).
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). This paper examines the transcriptionally engaged polymerase across the genome in human cells, and demonstrates a 59-end enrichment in engaged Pol II at many genes.
Boehm, A. K., Saunders, A., Werner, J. & Lis, J. T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 23, 7628–7637 (2003).
Peterson, C. L. & Workman, J. L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10, 187–192 (2000).
Larschan, E. & Winston, F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15, 1946–1956 (2001).
Ahn, S. H., Kim, M. & Buratowski, S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 39 end processing. Mol. Cell 13, 67–76 (2004).
Stargell, L. A. & Struhl, K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 12, 311–315 (1996).
Esnault, C. et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31, 337–346 (2008).
Kim, Y. J., Bjorklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 (1994).
Vermeulen, M. et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007).
Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Sakurai, H. & Fukasawa, T. Functional connections between mediator components and general transcription factors of Saccharomyces cerevisiae . J. Biol. Chem. 275, 37251–37256 (2000).
Liu, Y. et al. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 24, 1721–1735 (2004).
Spilianakis, C. et al. CIITA regulates transcription onset via Ser5-phosphorylation of RNA Pol II. EMBO J. 22, 5125–5136 (2003).
Core, L. J. & Lis, J. T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319, 1791–1792 (2008).
Ni, Z. et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo . Mol. Cell. Biol. 28, 1161–1170 (2008).
Chao, S. H. & Price, D. H. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo . J. Biol. Chem. 276, 31793–31799 (2001).
Eberhardy, S. R. & Farnham, P. J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 277, 40156–40162 (2002).
Wittmann, B. M., Fujinaga, K., Deng, H., Ogba, N. & Montano, M. M. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor α and transcriptional elongation factor cyclin T1. Oncogene 24, 5576–5588 (2005).
Nowak, D. E. et al. RelA Ser276 phosphorylation is required for activation of a subset of NF-κB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol. Cell. Biol. 28, 3623–3638 (2008).
Peterlin, B. M. & Price, D. H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006).
Mavrich, T. N. et al. Nucleosome organization in the Drosophila genome. Nature 453, 358–362 (2008).
Moore, M. J. & Proudfoot, N. J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 (2009).
Yudkovsky, N., Ranish, J. A. & Hahn, S. A transcription reinitiation intermediate that is stabilized by activator. Nature 408, 225–229 (2000).
Blau, J. et al. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16, 2044–2055 (1996).
Petesch, S. J. & Lis, J. T. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84 (2008).
Herschlag, D. & Johnson, F. B. Synergism in transcriptional activation: a kinetic view. Genes Dev. 7, 173–179 (1993). The paper proposes a role for kinetic synergism in the mechanism of transcriptional activation.
Boettiger A. N. & Levine, M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473 (2009).
Mason, P. B. & Struhl, K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo . Mol. Cell 17, 831–840 (2005).
Dejardin, J. & Kingston, R. E. Purification of proteins associated with specific genomic loci. Cell 136, 175–186 (2009).
Seila, A. C. et al. Divergent transcription from active promoters. Science 322, 1849–1851 (2008).
Preker, P. et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008).
Wuarin, J. & Schibler, U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 14, 7219–7225 (1994).
Bhaumik, S. R., Raha, T., Aiello, D. P. & Green, M. R. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18, 333–343 (2004).
Patterson, G. H. & Lippincott-Schwartz, J. Selective photolabeling of proteins using photoactivatable GFP. Methods 32, 445–450 (2004).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Yao, J., Munson, K. M., Webb, W. W. & Lis, J. T. Dynamics of heat shock factor association with native gene loci in living cells. Nature 442, 1050–1053 (2006).
Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004).
Chen, H.-T., Warfield, L. & Hahn, S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nature Struct. Mol. Biol. 14, 696–703 (2007).
Shi, H., Hoffman, B. E. & Lis, J. T. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA 96, 10033–10038 (1999).
Boeger, H., Griesenbeck, J., Strattan, J. S. & Kornberg, R. D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11, 1587–1598 (2003).
Svaren, J. & Hörz, W. Transcription factors vs nucleosomes: regulation of the PH05 promoter in yeast. Trends Biochem. Sci. 22, 93–97 (1997).
O'Neill, E. M., Kaffman, A., Jolly, E. R. & O'Shea, E. K. Regulation of PHO4 nuclear localization by the PHO80−PHO85 cyclin–CDK complex. Science 271, 209–212 (1996).
Barbaric, S., Munsterkotter, M., Goding, C. & Horz, W. Cooperative Pho2–Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol. 18, 2629–2639 (1998).
Svaren, J., Schmitz, J. & Horz, W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 13, 4856–4862 (1994).
Reinke, H. & Hörz, W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11, 1599–1607 (2003).
Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R. & O'Shea, E. K. Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116 (2003).
Korber, P. et al. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 281, 5539–5545 (2006).
Adkins, M. W., Howar, S. R. & Tyler, J. K. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14, 657–666 (2004).
Barbaric, S. et al. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo . J. Biol. Chem. 282, 27610–27621 (2007).
Lam, F. H., Steger, D. J. & O'Shea, E. K. Chromatin decouples promoter threshold from dynamic range. Nature 453, 246–250 (2008).
Wu, C. The 59 ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286, 854–860 (1980).
Lis, J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63, 347–356 (1998).
Lee, H., Kraus, K. W., Wolfner, M. F. & Lis, J. T. DNA sequence requirements for generating paused polymerase at the start of hsp70 . Genes Dev. 6, 284–295 (1992).
Wang, Y. V., Tang, H. & Gilmour, D. S. Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element. Mol. Cell. Biol. 25, 3543–3552 (2005).
Lee, C. et al. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila . Mol. Cell. Biol. 28, 3290–3300 (2008).
Tsukiyama, T., Becker, P. B. & Wu, C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367, 525–532 (1994).
Wu, C. H. et al. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 33, 1269–1279 (2005).
Wu, C.-H. et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila . Genes Dev. 17, 1402–1414 (2003).
Schwartz, B. E., Larochelle, S., Suter, B. & Lis, J. T. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo . Mol. Cell. Biol. 23, 6876–6886 (2003).
Ardehali, M. B. et al. Spt6 enhances the elongation rate of RNA polymerase II in vivo . EMBO J. 28, 1067–1077 (2009).
Lis, J. T., Mason, P., Peng, J., Price, D. H. & Werner, J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14, 792–803 (2000).
Renner, D. B., Yamaguchi, Y., Wada, T., Handa, H. & Price, D. H. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276, 42601–42609 (2001).
Ni, Z., Schwartz, B. E., Werner, J., Suarez, J. R. & Lis, J. T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13, 55–65 (2004).
Adelman, K. et al. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17, 103–112 (2005).
Acknowledgements
We thank members of the Lis laboratory for discussions and critical reading of the manuscript. Work in our laboratory is supported by US National Institutes of Health grant GM25232.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints.
Correspondence should be addressed to J.T.L. (jtl10@cornell.edu).
Rights and permissions
About this article
Cite this article
Fuda, N., Ardehali, M. & Lis, J. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192 (2009). https://doi.org/10.1038/nature08449
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08449
This article is cited by
-
Molecular mechanisms by which the HIV-1 latent reservoir is established and therapeutic strategies for its elimination
Archives of Virology (2023)
-
G-quadruplexes sense natural porphyrin metabolites for regulation of gene transcription and chromatin landscapes
Genome Biology (2022)
-
Mechanisms of enhancer action: the known and the unknown
Genome Biology (2021)
-
Molecular determinants of disease severity in urinary tract infection
Nature Reviews Urology (2021)
-
MYC protein interactors in gene transcription and cancer
Nature Reviews Cancer (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.