Abstract
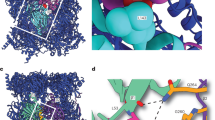
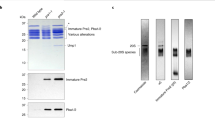
Substrates of the proteasome are recognized and unfolded by the regulatory particle, and then translocated into the core particle (CP) to be degraded1. A hetero-hexameric ATPase ring, containing subunits Rpt1-6, is situated within the base subassembly of the regulatory particle1. The ATPase ring sits atop the CP, with the Rpt carboxy termini inserted into pockets in the CP2,3,4,5,6. Here we identify a previously unknown function of the Rpt proteins in proteasome biogenesis through deleting the C-terminal residue from each Rpt in the yeast Saccharomyces cerevisiae. Our results indicate that assembly of the hexameric ATPase ring is templated on the CP. We have also identified an apparent intermediate in base assembly, BP1, which contains Rpn1, three Rpts and Hsm3, a chaperone for base assembly. The Rpt proteins with the strongest assembly phenotypes, Rpt4 and Rpt6, were absent from BP1. We propose that Rpt4 and Rpt6 form a nucleating complex to initiate base assembly, and that this complex is subsequently joined by BP1 to complete the Rpt ring. Our studies show that assembly of the proteasome base is a rapid yet highly orchestrated process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009)
Forster, A., Masters, E. I., Whitby, F. G., Robinson, H. & Hill, C. P. The 1.9 Å structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 18, 589–599 (2005)
Rabl, J. et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 (2008)
Smith, D. M. et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007)
Whitby, F. G. et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408, 115–120 (2000)
Gillette, T. G., Kumar, B., Thompson, D., Slaughter, C. A. & Demartino, G. N. Differential roles of the C-termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26s proteasome. J. Biol. Chem. 283, 31813–31822 (2008)
Borissenko, L. & Groll, M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 107, 687–717 (2007)
Mannhaupt, G., Schnall, R., Karpov, V., Vetter, I. & Feldmann, H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450, 27–34 (1999)
Xie, Y. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl Acad. Sci. USA 98, 3056–3061 (2001)
Isono, E. et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell 18, 569–580 (2007)
Kusmierczyk, A. R., Kunjappu, M. J., Funakoshi, M. & Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nature Struct. Mol. Biol. 15, 237–244 (2008)
Le Tallec, B., Barrault, M. B., Guerois, R., Carre, T. & Peyroche, A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell 33, 389–399 (2009)
Saeki, Y., Toh-e, A. & Yokosawa, H. Rapid isolation and characterization of the yeast proteasome regulatory complex. Biochem. Biophys. Res. Commun. 273, 509–515 (2000)
Leggett, D. S., Glickman, M. H. & Finley, D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 (2005)
Glickman, M. H. et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 (1998)
Roelofs, J. et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 10.1038/nature08063 (this issue)
Nakamura, Y. et al. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 359, 503–509 (2007)
Dawson, S. et al. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J. Biol. Chem. 277, 10893–10902 (2002)
Kress, W., Mutschler, H. & Weber-Ban, E. Assembly pathway of an AAA+ protein: tracking ClpA and ClpAP complex formation in real time. Biochemistry 46, 6183–6193 (2007)
Richmond, C., Gorbea, C. & Rechsteiner, M. Specific interactions between ATPase subunits of the 26 S protease. J. Biol. Chem. 272, 13403–13411 (1997)
Hartmann-Petersen, R., Tanaka, K. & Hendil, K. B. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 386, 89–94 (2001)
Gorbea, C., Taillandier, D. & Rechsteiner, M. Mapping subunit contacts in the regulatory complex of the 26S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem. 275, 875–882 (2000)
Rosenzweig, R., Osmulski, P. A., Gaczynska, M. & Glickman, M. H. The central unit within the 19S regulatory particle of the proteasome. Nature Struct. Mol. Biol. 15, 573–580 (2008)
Guthrie, C. & Fink, G. R. Guide to Yeast Genetics and Molecular Biology. (Academic Press, 1991)
Verma, R. et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 (2000)
Leggett, D. S. et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 (2002)
Li, X., Kusmierczyk, A. R., Wong, P., Emili, A. & Hochstrasser, M. Beta-subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 26, 2339–2349 (2007)
Kleijnen, M. F. et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nature Struct. Mol. Biol. 14, 1180–1188 (2007)
Hanna, J. et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 (2006)
Peng, J. & Gygi, S. P. Proteomics: the move to mixtures. J. Mass Spectrom. 36, 1083–1091 (2001)
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994)
Acknowledgements
We thank J. D. Orth for discussion and technical assistance, and C. Hill for discussions. We also thank J. W. Hanna, S. Elsasser and all members of Finley laboratory for comments on the manuscript. The work was supported by grants from US National Institutes of Health (NIH) to D.F. (GM043601) and S.P.G. (GM67945), an EMBO long-term fellowship to J.R. and an NIH NRSA postdoctoral fellowship (5F32GM75737-2) to S.P.
Author Contributions S.P. conducted all experiments. J.R. and D.F. contributed to designing experiments. W.K. performed mass spectrometry. S.P. and D.F. wrote the paper. All authors provided comments.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-5 with Legends, Supplementary Tables 1-2 and Supplementary References. (PDF 2487 kb)
Rights and permissions
About this article
Cite this article
Park, S., Roelofs, J., Kim, W. et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459, 866–870 (2009). https://doi.org/10.1038/nature08065
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08065
This article is cited by
-
Proteasome regulation by reversible tyrosine phosphorylation at the membrane
Oncogene (2021)
-
Regulation of proteasome assembly and activity in health and disease
Nature Reviews Molecular Cell Biology (2018)
-
Proteasome dysregulation in human cancer: implications for clinical therapies
Cancer and Metastasis Reviews (2017)
-
Reversible phosphorylation of the 26S proteasome
Protein & Cell (2017)
-
Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.