Abstract
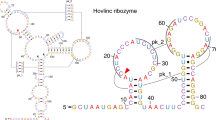
Structured RNAs embedded in the untranslated regions (UTRs) of messenger RNAs can regulate gene expression. In bacteria, control of a metabolite gene is mediated by the self-cleaving activity of a ribozyme embedded in its 5′ UTR1. This discovery has raised the question of whether gene-regulating ribozymes also exist in eukaryotic mRNAs. Here we show that highly active hammerhead ribozymes2,3 are present in the 3′ UTRs of rodent C-type lectin type II (Clec2) genes4,5,6,7. Using a hammerhead RNA motif search with relaxed delimitation of the non-conserved regions, we detected ribozyme sequences in which the invariant regions, in contrast to the previously identified continuous hammerheads8,9,10, occur as two fragments separated by hundreds of nucleotides. Notably, a fragment pair can assemble to form an active hammerhead ribozyme structure between the translation termination and the polyadenylation signals within the 3′ UTR. We demonstrate that this hammerhead structure can self-cleave both in vitro and in vivo, and is able to reduce protein expression in mouse cells. These results indicate that an unrecognized mechanism of post-transcriptional gene regulation involving association of discontinuous ribozyme sequences within an mRNA may be modulating the expression of several CLEC2 proteins that function in bone remodelling and the immune response of several mammals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Winkler, W. C. et al. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004)
Khvorova, A., Lescoute, A., Westhof, E. & Jayasena, S. D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nature Struct. Biol. 10, 708–712 (2003)
Martick, M. & Scott, W. G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320 (2006)
Zhou, H. et al. Osteoclast inhibitory lectin, a family of new osteoclast inhibitors. J. Biol. Chem. 277, 48808–48815 (2002)
Plougastel, B., Dubbelde, C. & Yokoyama, W. M. Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69 . Immunogenetics 53, 209–214 (2001)
Carlyle, J. R. et al. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc. Natl Acad. Sci. USA 101, 3527–3532 (2004)
Hao, L., Klein, J. & Nei, M. Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc. Natl Acad. Sci. USA 103, 3192–3197 (2006)
Przybilski, R. et al. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana . Plant Cell 17, 1877–1885 (2005)
Rojas, A. A. et al. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 28, 4037–4043 (2000)
Flores, R. et al. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 341, 540–552 (2001)
Gautheret, D., Major, F. & Cedergren, R. Pattern searching/alignment with RNA primary and secondary structures: an effective descriptor for tRNA. Comput. Appl. Biosci. 6, 325–331 (1990)
Eddy, S. R. RNABOB: a program to search for RNA secondary structure motifs in sequence databases. 〈http://selab.janelia.org/software.html〉 (2008)
Zhou, H. et al. A novel osteoblast-derived C-type lectin that inhibits osteoclast formation. J. Biol. Chem. 276, 14916–14923 (2001)
Carlyle, J. R. et al. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J. Immunol. 176, 7511–7524 (2006)
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002)
Salehi-Ashtiani, K., Luptak, A., Litovchick, A. & Szostak, J. W. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313, 1788–1792 (2006)
Teixeira, A. et al. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature 432, 526–530 (2004)
Ferbeyre, G., Smith, J. M. & Cedergren, R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell. Biol. 18, 3880–3888 (1998)
Serganov, A. & Patel, D. J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nature Rev. Genet. 8, 776–790 (2007)
Canny, M. D. et al. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 126, 10848–10849 (2004)
Osborne, E. M., Schaak, J. E. & Derose, V. J. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA 11, 187–196 (2005)
Martick, M. Structural Study of how the Hammerhead Ribozyme Solves the Problem of Catalysis. PhD thesis, Univ. California, 〈http://www.proquest.com/〉 (2007)
Yen, L. et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431, 471–476 (2004)
Meaux, S. & Van Hoof, A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA 12, 1323–1337 (2006)
Lockyer, A. E. et al. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology 126, 203–224 (2003)
Garneau, N. L., Wilusz, J. & Wilusz, C. J. The highways and byways of mRNA decay. Nature Rev. Mol. Cell Biol. 8, 113–126 (2007)
Kent, W. J. BLAT–the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002)
Stern, S., Moazed, D. & Noller, H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164, 481–489 (1988)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Acknowledgements
We thank M. Hall and K. Chakrabarti for assistance with cell culture, members of Haussler and Ares laboratories for sharing tissue culture space, M. Robertson for the transcriptase and A. Zahler for discussion. This work was supported by the National Institutes of Health grant R01043393.
Author Contributions L.H.H. and M.M. did the sequence searches, designed the study, performed the experiments, analysed the data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information 1
The file contains Supplementary Figures 1-3 and Legends and additional references. (PDF 296 kb)
Rights and permissions
About this article
Cite this article
Martick, M., Horan, L., Noller, H. et al. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454, 899–902 (2008). https://doi.org/10.1038/nature07117
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07117
This article is cited by
-
Zn2+-dependent DNAzymes that cleave all combinations of ribonucleotides
Communications Biology (2021)
-
Hunting for human ribozymes
Nature Chemical Biology (2021)
-
Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs
Genome Biology (2016)
-
RNA motif search with data-driven element ordering
BMC Bioinformatics (2016)
-
Sweet complementarity: the functional pairing of glycans with lectins
Cellular and Molecular Life Sciences (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.