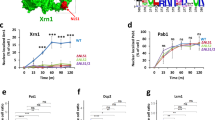
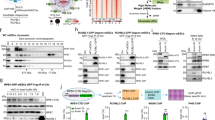
Abstract
Phosphoinositides are a family of lipid signalling molecules that regulate many cellular functions in eukaryotes. Phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P2), the central component in the phosphoinositide signalling circuitry, is generated primarily by type I phosphatidylinositol 4-phosphate 5-kinases (PIPKIα, PIPKIβ and PIPKIγ)1. In addition to functions in the cytosol, phosphoinositides are present in the nucleus2,3, where they modulate several functions4,5,6; however, the mechanism by which they directly regulate nuclear functions remains unknown. PIPKIs regulate cellular functions through interactions with protein partners, often PtdIns4,5P2 effectors, that target PIPKIs to discrete subcellular compartments, resulting in the spatial and temporal generation of PtdIns4,5P2 required for the regulation of specific signalling pathways1,7. Therefore, to determine roles for nuclear PtdIns4,5P2 we set out to identify proteins that interacted with the nuclear PIPK, PIPKIα. Here we show that PIPKIα co-localizes at nuclear speckles and interacts with a newly identified non-canonical poly(A) polymerase, which we have termed Star-PAP (nuclear speckle targeted PIPKIα regulated-poly(A) polymerase) and that the activity of Star-PAP can be specifically regulated by PtdIns4,5P2. Star-PAP and PIPKIα function together in a complex to control the expression of select mRNAs, including the transcript encoding the key cytoprotective enzyme haem oxygenase-1 (refs 8, 9) and other oxidative stress response genes by regulating the 3′-end formation of their mRNAs. Taken together, the data demonstrate a model by which phosphoinositide signalling works in tandem with complement pathways to regulate the activity of Star-PAP and the subsequent biosynthesis of its target mRNA. The results reveal a mechanism for the integration of nuclear phosphoinositide signals and a method for regulating gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
The Star-PAP sequence is deposited in the NCBI Library under accession number NP_073741. The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE9361.
References
Heck, J. N. et al. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit. Rev. Biochem. Mol. Biol. 42, 15–39 (2007)
Gonzales, M. L. & Anderson, R. A. Nuclear phosphoinositide kinases and inositol phospholipids. J. Cell. Biochem. 97, 252–260 (2006)
Boronenkov, I. V., Loijens, J. C., Umeda, M. & Anderson, R. A. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 9, 3547–3560 (1998)
Zhao, K. et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 (1998)
York, J. D., Odom, A. R., Murphy, R., Ives, E. B. & Wente, S. R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285, 96–100 (1999)
Osborne, S. L., Thomas, C. L., Gschmeissner, S. & Schiavo, G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 114, 2501–2511 (2001)
Doughman, R. L., Firestone, A. J. & Anderson, R. A. Phosphatidylinositol phosphate kinases put PI4,5P2 in its place. J. Membr. Biol. 194, 77–89 (2003)
Keum, Y. S. et al. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm. Res. 23, 2586–2594 (2006)
Duckers, H. J. et al. Heme oxygenase-1 protects against vascular constriction and proliferation. Nature Med. 7, 693–698 (2001)
Raabe, T., Bollum, F. J. & Manley, J. L. Primary structure and expression of bovine poly(A) polymerase. Nature 353, 229–234 (1991)
Rouhana, L. et al. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11, 1117–1130 (2005)
Cocco, L., Manzoli, L., Barnabei, O. & Martelli, A. M. Significance of subnuclear localization of key players of inositol lipid cycle. Adv. Enzyme Regul. 44, 51–60 (2004)
Kyriakopoulou, C. B., Nordvarg, H. & Virtanen, A. A novel nuclear human poly(A) polymerase (PAP), PAPγ. J. Biol. Chem. 276, 33504–33511 (2001)
Martin, G. & Keller, W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 15, 2593–2603 (1996)
Trippe, R. et al. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12, 1494–1504 (2006)
Rissland, O. S., Mikulasova, A. & Norbury, C. J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 27, 3612–3624 (2007)
LaCava, J. et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 (2005)
Colgan, D. F. & Manley, J. L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11, 2755–2766 (1997)
Wang, L., Eckmann, C. R., Kadyk, L. C., Wickens, M. & Kimble, J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312–316 (2002)
Takagaki, Y. & Manley, J. L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20, 1515–1525 (2000)
Hirose, Y. & Manley, J. L. RNA polymerase II and the integration of nuclear events. Genes Dev. 14, 1415–1429 (2000)
Mandel, C. R. et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444, 953–956 (2006)
Shatkin, A. J. & Manley, J. L. The ends of the affair: capping and polyadenylation. Nature Struct. Biol. 7, 838–842 (2000)
Nagaoka, K., Suzuki, T., Kawano, T., Imakawa, K. & Sakai, S. Stability of casein mRNA is ensured by structural interactions between the 3′-untranslated region and poly(A) tail via the HuR and poly(A)-binding protein complex. Biochim. Biophys. Acta 1759, 132–140 (2006)
Gilbert, C., Kristjuhan, A., Winkler, G. S. & Svejstrup, J. Q. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14, 457–464 (2004)
Christofori, G. & Keller, W. 3′ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell 54, 875–889 (1988)
Zhao, J., Hyman, L. & Moore, C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 (1999)
Stevenson, A. L. & Norbury, C. J. The Cid1 family of non-canonical poly(A) polymerases. Yeast 23, 991–1000 (2006)
Kwak, J. E. & Wickens, M. A family of poly(U) polymerases. RNA 13, 860–867 (2007)
Ding, J. et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 13, 1102–1115 (1999)
Ling, K., Doughman, R. L., Firestone, A. J., Bunce, M. W. & Anderson, R. A. Type Iγ phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420, 89–93 (2002)
Kendziorski, C. M., Newton, M. A., Lan, H. & Gould, M. N. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat. Med. 22, 3899–3914 (2003)
Acknowledgements
We thank J. L. Manley for the gift of antibodies against CPSF-73, CstF-64 and PAPα; M. Wickens for the generous gift of the PAPα construct and for advice and discussion; D. Brow, S. Miyamoto, D. Wassarman and R. Tibbetts for reading the manuscript and for comments; and C. Song for technical assistance in the early parts of the project. R.A.A. is supported by grants from the National Institutes of Health (NIH). M.L.G., D.L.M. and C.S. were supported by the American Heart Association. C.A.B. is supported by the National Research Service Award. D.L.M. and M.L.G. received Research Training Grant support from the NIH.
Author Contributions D.L.M. contributed to Figs 1a, d, f, 2, 3a, b, 4g–k and Supplementary Figs 1, 2, 4–7. M.L.G. contributed to Figs 1a, 3c, f, 4a–f and Supplementary Fig. 1. C.A.B. contributed to Fig. 3d, e, Supplementary Fig. 1 and Supplementary Tables 1 and 2. C.S. contributed to Fig. 1a, c, e and Supplementary Figs 3 and 6. P.W. and C.K. analysed the microarray data. R.A.A. directed the experimental approach and project. D.L.M., M.L.G., C.A.B. and R.A.A. analysed and interpreted the experiments, and conceptualized and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Methods, Supplementary Figures 1-7 with Legends and Supplementary Tables 1-2. (PDF 11680 kb)
Rights and permissions
About this article
Cite this article
Mellman, D., Gonzales, M., Song, C. et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451, 1013–1017 (2008). https://doi.org/10.1038/nature06666
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06666
This article is cited by
-
Nuclear speckleopathies: developmental disorders caused by variants in genes encoding nuclear speckle proteins
Human Genetics (2023)
-
A p53–phosphoinositide signalosome regulates nuclear AKT activation
Nature Cell Biology (2022)
-
A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing
Nature Reviews Molecular Cell Biology (2020)
-
Genome wide association and gene enrichment analysis reveal membrane anchoring and structural proteins associated with meat quality in beef
BMC Genomics (2019)
-
A nuclear phosphoinositide kinase complex regulates p53
Nature Cell Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.