Abstract
The naturally occurring regulatory T cell (Tr) is the pivotal cell type that maintains self-tolerance and exerts active immune suppression. The development and function of Tr cells is controlled by Foxp3 (refs 1, 2), a lack of which results in loss of Tr cells and massive multi-organ autoimmunity in scurfy mice and IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked) patients3,4. It is generally thought that, through a binary mechanism, Foxp3 expression serves as an on-and-off switch to regulate positively the physiology of Tr cells; however, emerging evidence associates decreased Foxp3 expression in Tr cells with various immune disorders5,6,7. We hypothesized that Foxp3 regulates Tr cell development and function in a dose-dependent, non-binary manner, and that decreased Foxp3 expression can cause immune disease. Here, by generating a mouse model in which endogenous Foxp3 gene expression is attenuated in Tr cells, we show that decreased Foxp3 expression results in the development of an aggressive autoimmune syndrome similar to that of scurfy mice, but does not affect thymic development, homeostatic expansion/maintenance or transforming-growth-factor-β-induced de novo generation of Foxp3-expressing cells. The immune-suppressive activities of T cells with attenuated Foxp3 expression were nearly abolished in vitro and in vivo, whereas their anergic properties in vitro were maintained. This was accompanied by decreased expression of Tr cell ‘signature genes’. Notably, T cells expressing decreased Foxp3 preferentially became T-helper 2 (Th2)-type effectors even in a Th1-polarizing environment. These cells instructed Th2 differentiation of conventional T cells, which contributed to the immune diseases observed in these mice. Thus, decreased Foxp3 expression causes immune disease by subverting the suppressive function of Tr cells and converting Tr cells into effector cells; these findings are important for understanding the regulation of Tr cell function and the aetiology of various human immune diseases.
Similar content being viewed by others
Main
Tr cells, a central component for immune suppression, are critical for establishing self-tolerance, controlling inflammatory responses and maintaining immune homeostasis8,9. Foxp3, an X-chromosome-linked factor that controls Tr cell development and function1,2, is generally thought to control positively the functions of Tr cells in a binary fashion, as Foxp3 expression is sufficient to specify immune-suppressive activities in conventional T cells1,2,10. Thus, current efforts are focused on associating abnormal numbers of Tr cells with immune disorders. However, the quality of Tr cells is also critical for their function11. We observed lowered levels of Foxp3 in intra-islet Tr cells compared with Tr cells from other peripheral lymphoid organs in diabetic NOD mice (Fig. 1a), whereas the frequencies of Foxp3-expressing Tr cells among different compartments were comparable (data not shown). However, such a specific decrease in Foxp3 expression was not observed in non-diabetes-prone C57BL/6 mice (Supplementary Fig. 1). We hypothesized that one of the quality control mechanisms for Tr cells is through tuning the expression levels of Foxp3, and that decreased Foxp3 expression can cause immune disease. To test this, we generated a mouse model, where attenuated expression of the endogenous Foxp3 gene was achieved by a targeted gene ‘knock-in’ approach, allowing us to investigate the effects of decreased Foxp3 expression on Tr cell function and to provide potential mechanistic explanations for the aetiologies of certain human immune disorders. In this model, a gene cassette co-expressing luciferase and enhanced green fluorescent protein (eGFP)—the translation of which was under the control of two tandem internal ribosomal entry sites (IRES)—was inserted into the 3′-untranslated region (UTR) of the endogenous Foxp3 locus of C57BL/6 mice to generate a Foxp3-IRES-luciferase-IRES-eGFP (FILIG) allele (Supplementary Fig. 2). Using a similar approach, we have previously generated a knock-in mouse model where Foxp3-expressing cells are marked by the co-expression of a monomeric red fluorescent protein (RFP)12. Such Foxp3-IRES-mRFP (FIR) mice can be used to isolate wild-type Tr cells based on RFP expression. In the following experiments, hemizygous male FIR mice (FIR/Y), heterozygous female FIR mice (FIR/+) or homozygous female FIR mice (FIR/FIR) and T cells from these mice were used as wild-type controls where appropriate.
a, Flow cytometry for Foxp3 expression in CD4+ T cells isolated from intra-islet lymphocytes (IIL; green line), and axillary (ALN), superficial-inguinal (ILN) and submandibular (SLN) lymph nodes (blue lines) from diabetic NOD mice. Mean fluorescence intensity (MFI) of Foxp3 staining among Foxp3+ cells is indicated. Data are mean ± s.d. of two samples with one sample combining lymphocytes from three mice. b, Size comparison of 2-month-old FIR/Y and FILIG/Y mice (left panel); sizes of submandibular (SLN), axillary (ALN) and superficial-inguinal (ILN) lymph nodes and spleens are also shown (right panel). c, Localization of luciferase-expressing cells in various organs in FILIG/+ and FILIG/Y mice. The colour scale with corresponding count units is shown. d, The percentage (highlighted in red) of GFP+ cells among CD4+ T cells in FILIG/Y and FILIG/+ mice and of RFP+ cells in FIR/Y and FIR/+ control mice is shown. Results representative of at least ten experiments are shown. e, Flow cytometry of Foxp3 in sorted GFP+ CD4+ T cells from FILIG/+ mice; GFP+ and GFP- CD4+ T cells from FILIG/Y mice; and RFP+ and RFP- CD4+ T cells from FIR/Y mice. Results representative of four experiments are shown. f, Surface expression of CD25, CTLA4 and GITR on GFP+ CD4+ T cells from FILIG/+ mice (blue lines); RFP+ CD4+ T cells from FIR/+ mice (red lines); and RFP– CD4+ T cells from FIR/FIR mice (green lines). Results are representative of three experiments. g, Relative mRNA levels of Foxp3, CD25, Ctla4 and Gitr in sorted GFP+ CD4+ T cells (grey bars) from FILIG/+ mice, and RFP+ (white bars) and RFP– (black bars) CD4+ T cells from FIR/FIR mice. Data are mean ± s.d. of combined results from three experiments. The differences observed among samples for different genes are statistically significant as determined by the ‘Statistical analysis’ section of the Methods.
FILIG mice were born at a mendelian ratio. Heterozygous FILIG female mice (FILIG/+) were fertile and phenotypically normal; however, hemizygous FILIG male mice (FILIG/Y) were barren and runted (Fig. 1b). Over 50% of FILIG/Y mice developed scaly skin (data not shown) and nearly all of them developed eyelid defects resembling blepharitis, a Th2 disorder, at around 4 weeks of age (Supplementary Fig. 3). By 3 months of age, all the FILIG/Y mice succumbed to an aggressive lymphoproliferative autoimmune syndrome, manifested by enlarged spleens and lymph nodes (Fig. 1b), infiltration of lymphocytes into non-lymphoid organs, drastically increased serum levels of auto-antibodies (Supplementary Fig. 4), and activated CD4+ and CD8+ T cells (Supplementary Fig. 5). Overall, FILIG/Y mice displayed phenotypes reminiscent of scurfy mice13 and T-cell-specific Foxp3 knockout mice1. To investigate whether transcription of the endogenous Foxp3 gene was abolished in FILIG mice, we first detected luciferase expression by live imagining. In FILIG/+ mice, cells expressing luciferase were concentrated in lymphoid organs (Fig. 1c). However, in FILIG/Y mice, these cells were detected in lymphoid as well as non-lymphoid organs (Fig. 1c), suggesting that the endogenous Foxp3 gene was transcribed in FILIG lymphocytes, and that FILIG lymphocytes infiltrated non-lymphoid organs in FILIG/Y mice. By flow cytometry, we detected GFP expression only in CD4+ T cells (Supplementary Fig. 6). Notably, among CD4+ T cells, there was a higher percentage of GFP+ cells in FILIG/Y mice compared with RFP+ cells in FIR/Y mice, whereas a lower percentage of GFP+ cells was detected in FILIG/+ mice compared with RFP+ cells in FIR/+ mice (Fig. 1d). By intracellular staining, we detected Foxp3 expression in GFP+ CD4+ T cells from FILIG/Y and FILIG/+ mice, but not in GFP- CD4+ T cells from FILIG/Y mice (Fig. 1e). Therefore, GFP expression reflected Foxp3 expression with high fidelity in FILIG mice. Compared with wild-type Tr cells, GFP+ cells from FILIG mice expressed 5–10-fold less Foxp3. Thus, although Foxp3 was expressed at decreased levels, FILIG mice generated Foxp3-expressing CD4+ T cells that were faithfully marked by GFP expression. Compared with wild-type Tr cells, the surface expression and messenger RNA levels of signature genes for Tr cells (such as CD25, Ctla4 and Gitr14,15,16) were decreased in GFP+ CD4+ T cells from FILIG/+ mice (Fig. 1f, g). The exact mechanism by which attenuated Foxp3 expression was achieved in FILIG CD4+ T cells is unclear. However, four AU-rich elements (ARE), the presence of which in the 3′-UTR of a gene is known to destabilize mRNA17, were found dispersed in the luciferase complementary DNA. Therefore, reduced Foxp3 expression in FILIG CD4+ T cells is probably due to mRNA destabilization caused by localization of the luciferase sequence in the 3′-UTR of the Foxp3 mRNA. As a consequence of random X-chromosome inactivation, the FILIG/+ female mice contain a mixture of Foxp3+ cells: those with wild-type levels of Foxp3 expression and those having low levels due to the gene knock-in approach; this mixture of cells probably accounts for lack of overt disease in these mice.
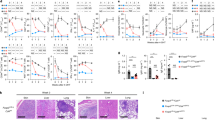
The FILIG model allowed us to investigate further which biological functions of Tr cells are altered owing to decreased Foxp3 expression. To assess whether attenuated Foxp3 expression affected Tr cell development, the percentages of Foxp3-expressing cells (Foxp3+) among CD4+ single positive thymocytes from 10- and 16-day-old FILIG/Y and FIR/Y mice were determined and compared, but no difference was observed (Fig. 2a). Whether homeostatic expansion/maintenance of Foxp3+ cells was affected by attenuated Foxp3 expression was addressed by transferring cell mixtures combining conventional CD4+ T cells (RFP-) from FIR/FIR mice with Foxp3+ (RFP+ or GFP+) CD4+ T cells from either FIR/+ or FILIG/+ mice into Rag1-/- mice. RFP+ and GFP+ CD4+ T cells in the recipient mice were detected 9 weeks after transfer (Supplementary Fig. 7); the percentage of RFP+ and GFP+ cells among transferred CD4+ T cells was comparable (Fig. 2b), suggesting that decreased Foxp3 expression did not result in intrinsic defects of the homeostatic expansion/maintenance of GFP+ FILIG cells. However, when RFP+ and GFP+ CD4+ T cells were co-transferred into the same hosts in the presence of conventional CD4+ T cells, GFP+ cells competed poorly with RFP+ cells (Fig. 2c), in agreement with the observation that a lower than expected percentage of GFP+ cells was found in the FILIG/+ mice. The fact that CD25 expression on GFP+ cells from FILIG/+ mice was decreased relative to that on RFP+ cells from FIR/+ mice could account for this phenomenon, as Tr cell maintenance is dependent on interleukin-2 (IL-2) signalling18,19,20. Extra-thymic generation of Foxp3+ T cells can be promoted in vitro by transforming growth factor-β (TGF-β)12,21,22. TGF-β induced de novo Foxp3 expression in FILIG CD4+ T cells to a similar extent as in FIR CD4+ T cells (Fig. 2d).
a, The percentage of Foxp3+ CD4+ single positive thymocytes in 10- and 16-day-old FIR/Y (white bars) and FILIG/Y mice (black bars) is shown. Data are mean ± s.d. of results from four mice for each time point. b, Sorted RFP- CD4+ T cells from FIR/FIR mice were mixed with either RFP+ CD4+ T cells from FIR/+ mice (white bars) or GFP+ CD4+ T cells from FILIG/+ mice (black bars) at a ratio of 2:1 and then transferred into female Rag1-/- mice. The percentage of Foxp3+ (RFP+ or GFP+) CD4+ T cells from different origins in different lymphoid organs was determined by flow cytometry 9 weeks later. PLN, peripheral lymph nodes and spleen. Data are mean ± s.d. of results from six mice of one experiment representative of two. c, RFP+ and GFP+ CD4+ T cells were sorted from FIR/+ and FILIG/+ mice respectively, and then combined at the indicated ratios. 1×105 cell mixtures and 2×105 RFP- CD4+ T cells sorted from FIR/FIR mice were co-transferred into Rag1-/- mice. The distribution of Foxp3+ FIR (RFP+) and Foxp3+ FILIG (GFP+) cells in the recipient mice was determined by flow cytometry 9 weeks later. Results representative of six mice are shown. d, CD4+CD25- T cells that were negative for RFP and GFP were sorted from FIR/+ and FILIG/+ mice respectively, and then activated in the presence of TGF-β1. Foxp3 expression was measured by the expression of RFP and GFP. Results representative of three experiments are shown.
In vitro, anergy and immune-suppressive activities are two defining properties for Tr cells that are thought to go hand-in-hand23. Notably, upon T-cell-receptor (TCR) stimulation in vitro, although GFP+ cells from FILIG mice remained anergic (Fig. 3a and Supplementary Fig. 8), their immune-suppressive activities were greatly impaired (Fig. 3b). Thus, anergy and immune suppression are two separable properties of Tr cells that are affected differentially by Foxp3 expression level. The immune-suppressive activities of Foxp3+ CD4+ T cells from FILIG mice were also abolished in vivo, because, unlike wild-type Tr cells, co-transferred GFP+ cells from FILIG/+ mice did not prevent conventional CD4+ T-cell-elicited weight loss (Fig. 3c) or the immune pathologies in the intestines and stomach (Fig. 3d) of Rag1–/– recipient mice. This was not due to defective migration of transferred cells. In fact, adoptively transferred GFP+ FILIG cells are found in lymphoid as well as non-lymphoid organs (Fig. 3e).
a, RFP+, GFP+, RFP-CD25- and GFP-CD25- CD4+ T cells were sorted from FIR/+ (white bars) and FILIG/+ (black bars) mice. Purified cells were activated in vitro, and cell proliferation was determined by a T-cell proliferation assay. Data are mean ± s.d. of triplicates done in one experiment representative of three. b, Suppression assay performed using sorted RFP+ CD4+ T cells from FIR/+ mice (blue line) or GFP+ CD4+ T cells from FILIG/+ mice (red line) as suppressor cells (S) and RFP- CD4+ T cells sorted from FIR/FIR mice as responder cells (R). Data are mean ± s.d. of triplicates done in one experiment representative of three. c, Rag1-/- mice were transferred with RFP- CD4+ T cells sorted from FIR/FIR mice alone (black line), or with T-cell mixtures containing one-third of RFP+ CD4+ T cells sorted from FIR/+ mice (blue line) or GFP+ CD4+ T cells sorted from FILIG/+ mice (red line) and two-thirds of RFP- CD4+ T cells sorted from FIR/FIR mice. The percentage of body weight change in the recipient mice was determined weekly for 9 weeks after transfer. Data are mean ± s.d. of six mice from one experiment representative of two. d, Intestines and stomach were removed from recipient mice at the end of the experiments described in c. Haematoxylin-and-eosin staining was performed to detect pathological changes in mice that received conventional CD4+ T cells together with RFP+ CD4+ T cells from FIR/+ (+FIR) or GFP+ CD4+ T cells from FILIG/+ (+FILIG) mice; or without Foxp3+ cells (-). e, At the end of experiments described in c, localization of transferred GFP+ FILIG cells in different organs (as indicated) was determined by live imaging. The colour scale with corresponding count units is shown. As a control, organs from a mouse that received RFP+ FIR cells are shown.
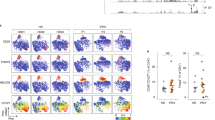
Loss of the suppressive activity of Tr cells could be sufficient to cause an aggressive autoimmune syndrome in FILIG/Y mice and the wasting disease observed in the aforementioned transfer model. Nevertheless, we further investigated whether Foxp3+ FILIG cells developed effector functions that could contribute to any of these immune disorders. On the basis of cytokine production profiles, three types of effector T cells, Th1, Th2 and Th17, have been described24,25. Although Tr cells bear self-reactive TCRs26,27, GFP+ cells from healthy FILIG/+ mice did not exhibit an effector cell phenotype (Fig. 4a), suggesting that they did not spontaneously activate and convert into effector cells when substantial numbers of wild-type Tr cells were present. Notably, however, a large portion of GFP+ cells from diseased FILIG/Y mice produced IL-4, whereas the percentage of cells expressing IL-2, interferon-γ (IFN-γ) or IL-17 was only modestly increased compared to wild-type cells (Fig. 4b). The percentage of IL-4-producing cells also preferentially increased in Foxp3- (GFP–) CD4+ T cells from FILIG/Y mice, consistent with the Th2 disorder observed in these mice. Cytokine mRNA levels were also determined and agreed with aforementioned results (Fig. 4c). In addition, compared with wild-type Tr cells, the percentage of IL-4-producing GFP+ FILIG cells showed an approximately 30-fold increase upon adoptive transfer, whereas that of IFN-γ-producing GFP+ FILIG cells showed only an approximately 3-fold increase (Fig. 4d). In some experiments, we also noticed that the percentage of IL-4- but not IFN-γ-producing cells was substantially increased even among transferred wild-type Foxp3+ FIR cells that expressed lower levels of Foxp3 (Supplementary Fig. 9), suggesting that IL-4 production can be induced in wild-type Tr cells expressing low levels of Foxp3 in vivo. To compare Foxp3+ FIR and FILIG CD4+ T cells directly in the same physiological environment, CD4+ T cells that were RFP+ from FIR/+ mice, GFP+ from healthy FILIG/+ mice, and RFP– from FIR/FIR mice, were mixed at a ratio of 1:10:20 and then transferred into Rag1–/– hosts. At the time of transfer, none of these cells was producing substantial amounts of effector cytokines (Fig. 4a and data not shown). Recipient mice developed wasting disease 9 weeks after transfer (data not shown), possibly owing to the fact that insufficient numbers of wild-type Tr cells were transferred. Compared with coexisting RFP+ wild-type Tr cells, the fraction of IL-4-producing cells among GFP+ FILIG cells increased greatly, whereas that of IFN-γ- or IL-17-producing cells did not change substantially, although coexisting Foxp3– cells produced large quantities of IFN-γ, thereby providing a Th1-polarizing condition (Fig. 4e). Moreover, GFP+ FILIG cells potently induced Th2 differentiation of conventional CD4+ T cells in vitro and in vivo, potentially through IL-4. Intriguingly, IL-4 production by GFP+ FILIG cells was not affected by the Th1-polarizing environments imposed by co-cultured cells (Supplementary Fig. 10).
a, b, The production of IL-2, IL-4, IL-17 and IFN-γ in RFP+ CD4+ T cells sorted from FIR/+ mice and in GFP+ CD4+ T cells sorted from FILIG/+ mice (a), or in RFP+ and RFP- CD4+ T cells from FIR/Y mice and GFP+ and GFP- CD4+ T cells from FILIG/Y mice (b) is shown. c, Relative mRNA levels of Ifng, Lta (lymphotoxin A), Ltb (lymphotoxin B), Il4, Il13, Il10, Il2 and Il17 among RFP+, RFP-, GFP+ and GFP– CD4+ T cells isolated from male FIR/Y and FILIG/Y mice are shown. Data are mean ± s.d. from two experiments. d, At the end of the experiments described in Fig. 3c, The expression of IL-4 and IFN-γ was examined in RFP+ FIR/+ and GFP+ FILIG/+ cells that were recovered from recipient mice. e, At the end of the experiments described in Fig. 2c (right panel), the expression of IL-4, IL-17 and IFN-γ was determined in co-existing RFP+ FIR/+ cells and GFP+ FILIG/+ cells as well as in RFP-GFP- FIR/FIR cells. All results are representative of at least three experiments unless stated otherwise.
Decreased Foxp3 expression is associated with human immune disorders5,6,7. Our genetic evidence convincingly shows that decreased Foxp3 expression can cause defective suppressive function of Tr cells and their conversion into effector cells, which contribute to rather than inhibit immune diseases. This provides an important mechanistic explanation for the aetiology of immunopathology in our mouse model and potentially various human immune diseases.
Methods
Mice and adoptive transfer assays
FIR, FILIG, NOD, C57BL/6 and Rag1–/– (C57BL/6 background) mice were kept under specific pathogen-free conditions in the animal care facility at Yale University. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Yale University. For adoptive transfer assays, conventional (RFP-) CD4+ T cells from FIR/FIR mice, RFP+ CD4+ T cells from FIR/+ mice or GFP+ CD4+ T cells from FILIG/+ mice were sorted by fluorescence-activated cell sorting (FACS). Sorted cells were either transferred alone or mixed at different ratios as elaborated in the text or figure legends. A total of 3×105 cells were transferred into female Rag1–/– mice via retro-orbital injection. Mice were weighed every week thereafter and killed 9–10 weeks after transfer.
Generation of FILIG mice
FILIG knock-in mice were generated according to the protocols described for generating FIR mice12. A gene cassette encoding IRES-luciferase-IRES-eGFP instead of IRES-RFP was inserted into an Ssp1 site. The floxed neomycin cassette was deleted in vitro by transfecting Cre-expressing plasmid into Bruce-4 embryonic stem cells originating from C57BL/6 mice.
Live imaging and histology
For live imaging analysis, mice were injected intraperitoneally with 3 mg luciferin (Xenogen) per mouse and then killed. Different lymphoid and non-lymphoid organs were surgically removed. Luciferase-expressing organs were visualized by IVIS Imaging System (Xenogen) as per the manufacturer’s protocols. For histology analysis, organs from mice were removed and fixed in S.T.F. fixatives (Streck) for 24 h. Preparation of the slides, sectioning and haematoxylin-and-eosin staining were performed by the Yale histology and pathology laboratory.
Statistical analysis
Data from at least three sets of samples were used for statistical analysis. Mean ± s.d. are shown. Statistical significance was calculated by Student’s t-test. A P-value of less than 0.05 was considered significant.
References
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature Immunol. 4, 330–336 (2003)
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003)
Wildin, R. S. et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nature Genet. 27, 18–20 (2001)
Brunkow, M. E. et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nature Genet. 27, 68–73 (2001)
Miura, Y. et al. Association of Foxp3 regulatory gene expression with graft-versus-host disease. Blood 104, 2187–2193 (2004)
Balandina, A., Lecart, S., Dartevelle, P., Saoudi, A. & Berrih-Aknin, S. Functional defect of regulatory CD4+CD25+ T cells in the thymus of patients with autoimmune myasthenia gravis. Blood 105, 735–741 (2005)
Huan, J. et al. Decreased FOXP3 levels in multiple sclerosis patients. J. Neurosci. Res. 81, 45–52 (2005)
Sakaguchi, S. Regulatory T cells: key controllers of immunologic self-tolerance. Cell 101, 455–458 (2000)
Shevach, E. M. Regulatory T cells in autoimmunity. Annu. Rev. Immunol. 18, 423–449 (2000)
Khattri, R., Cox, T., Yasayko, S. A. & Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nature Immunol. 4, 337–342 (2003)
Fontenot, J. D. & Rudensky, A. Y. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nature Immunol. 6, 331–337 (2005)
Wan, Y. Y. & Flavell, R. A. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl Acad. Sci. USA 102, 5126–5131 (2005)
Lyon, M. F., Peters, J., Glenister, P. H., Ball, S. & Wright, E. The scurfy mouse mutant has previously unrecognized hematological abnormalities and resembles Wiskott-Aldrich syndrome. Proc. Natl Acad. Sci. USA 87, 2433–2437 (1990)
Sakaguchi, S., Sakaguchi, N., Asano, M., Itoh, M. & Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor α-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155, 1151–1164 (1995)
Takahashi, T. et al. Immunologic self-tolerance maintained by CD25+CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J. Exp. Med. 192, 303–310 (2000)
Ono, M., Shimizu, J., Miyachi, Y. & Sakaguchi, S. Control of autoimmune myocarditis and multiorgan inflammation by glucocorticoid-induced TNF receptor family-related proteinhigh, Foxp3-expressing CD25+ and CD25- regulatory T cells. J. Immunol. 176, 4748–4756 (2006)
Chen, C. Y. & Shyu, A. B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470 (1995)
Furtado, G. C., Curotto de Lafaille, M. A., Kutchukhidze, N. & Lafaille, J. J. Interleukin 2 signaling is required for CD4+ regulatory T cell function. J. Exp. Med. 196, 851–857 (2002)
Fontenot, J. D., Rasmussen, J. P., Gavin, M. A. & Rudensky, A. Y. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nature Immunol. 6, 1142–1151 (2005)
D’Cruz, L. M. & Klein, L. Development and function of agonist-induced CD25+Foxp3+ regulatory T cells in the absence of interleukin 2 signaling. Nature Immunol. 6, 1152–1159 (2005)
Chen, W. et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003)
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector Th17 and regulatory T cells. Nature 441, 235–238 (2006)
Sakaguchi, S. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 22, 531–562 (2004)
Mosmann, T. R. & Coffman, R. L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 7, 145–173 (1989)
Park, H. et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nature Immunol. 6, 1133–1141 (2005)
Hsieh, C. S., Zheng, Y., Liang, Y., Fontenot, J. D. & Rudensky, A. Y. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nature Immunol. 7, 401–410 (2006)
Hsieh, C. S. et al. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity 21, 267–277 (2004)
Acknowledgements
This research is supported by the NIH, American Diabetes Association and Howard Hughes Medical Institute. R.A.F. is an investigator of the Howard Hughes Medical Institute. Y.Y.W. is supported by a postdoctoral fellowship from the Cancer Research Institute. We thank L. Evangelisti, C. Hughes and J. Stein for assisting with the generation of FILIG mice. We are grateful to E. Eynon and L. Zenewicz for critical reading and helpful comments. We also thank F. Manzo and R. Champion for secretarial assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
This file contains Supplementary Methods with more detailed description of experimental procedures described in the manuscript and Supplementary Figures S1-S10. (PDF 679 kb)
Rights and permissions
About this article
Cite this article
Wan, Y., Flavell, R. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770 (2007). https://doi.org/10.1038/nature05479
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05479
This article is cited by
-
The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases
Nature Reviews Immunology (2024)
-
Regulatory T cells in the face of the intestinal microbiota
Nature Reviews Immunology (2023)
-
Longitudinal monitoring of mRNA levels of regulatory T cell biomarkers by using non-invasive strategies to predict outcome in renal transplantation
BMC Nephrology (2022)
-
Characterization of regulatory T cells in SARS-CoV-2 infected hemodialysis patients: relation to clinical and radiological severity
BMC Nephrology (2022)
-
Dynamic transcriptional activity and chromatin remodeling of regulatory T cells after varied duration of interleukin-2 receptor signaling
Nature Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.