Abstract
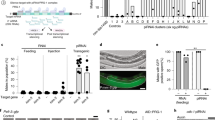
Paramutation is a heritable epigenetic modification induced in plants by cross-talk between allelic loci. Here we report a similar modification of the mouse Kit gene in the progeny of heterozygotes with the null mutant Kittm1Alf (a lacZ insertion). In spite of a homozygous wild-type genotype, their offspring maintain, to a variable extent, the white spots characteristic of Kit mutant animals. Efficiently inherited from either male or female parents, the modified phenotype results from a decrease in Kit messenger RNA levels with the accumulation of non-polyadenylated RNA molecules of abnormal sizes. Sustained transcriptional activity at the postmeiotic stages—at which time the gene is normally silent—leads to the accumulation of RNA in spermatozoa. Microinjection into fertilized eggs either of total RNA from Kittm1Alf/+ heterozygotes or of Kit-specific microRNAs induced a heritable white tail phenotype. Our results identify an unexpected mode of epigenetic inheritance associated with the zygotic transfer of RNA molecules.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brink, R. A. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41, 872–879 (1956)
Chandler, V. L. & Stam, M. Chromatin conversations: mechanisms and implications of paramutation. Nature Rev. Genet. 5, 532–544 (2004)
Rassoulzadegan, M., Magliano, M. & Cuzin, F. Transvection effects involving DNA methylation during meiosis in the mouse. EMBO J. 21, 4404–4450 (2002)
Herman, H. et al. Trans allele methylation and paramutation-like effects in mice. Nature Genet. 34, 199–202 (2003)
Bernex, F. et al. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development 122, 3023–3033 (1996)
Yasuda, H., Galli, S. J. & Geissler, E. N. Cloning and functional analysis of the mouse c-kit promoter. Biochem. Biophys. Res. Commun. 191, 893–901 (1993)
Olek, A., Oswald, J. & Walter, J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24, 5064–5066 (1996)
Lachner, M. & Jenuwein, T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14, 286–298 (2002)
Kluppel, M., Nagle, D. L., Bucan, M. & Bernstein, A. Long-range genomic rearrangements upstream of Kit dysregulate the developmental pattern of Kit expression in W57 and Wbanded mice and interfere with distinct steps in melanocyte development. Development 124, 65–77 (1997)
Manova, K., Nocka, K., Besmer, P. & Bachvarova, R. F. Gonadal expression of c-kit encoded at the W locus of the mouse. Development 110, 1057–1069 (1990)
Motro, B., van der Kooy, D., Rossant, J., Reith, A. & Bernstein, A. Contiguous patterns of c-kit and steel expression: analysis of mutations at the W and Sl loci. Development 113, 1207–1221 (1991)
Sorrentino, V., Giorgi, M., Geremia, R., Besmer, P. & Rossi, P. Expression of the c-kit proto-oncogene in the murine male germ cells. Oncogene 6, 149–151 (1991)
Vincent, S. et al. Stage-specific expression of the Kit receptor and its ligand (KL) during male gametogenesis in the mouse: a Kit–KL interaction critical for meiosis. Development 125, 4585–4593 (1998)
Albanesi, C. et al. A cell- and developmental stage-specific promoter drives the expression of a truncated c-kit protein during mouse spermatid elongation. Development 122, 1291–1302 (1996)
Turner, J. M. et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nature Genet. 37, 41–47 (2005)
Krawetz, S. A. Paternal contribution: new insights and future challenges. Nature Rev. Genet. 6, 633–642 (2005)
Darzynkiewicz, Z. Differential staining of DNA and RNA in intact cells and isolated cell nuclei with acridine orange. Methods Cell Biol. 33, 285–298 (1990)
Bernhard, W. A new staining procedure for electron microscopical cytology. J. Ultrastruct. Res. 27, 250–265 (1969)
Biggiogera, M. & Fakan, S. Fine structural specific visualization of RNA on ultrathin sections. J. Histochem. Cytochem. 46, 389–395 (1998)
Bernstein, E. & Allis, C. D. RNA meets chromatin. Genes Dev. 19, 1635–1655 (2005)
Hogan, B., Beddington, R., Costantini, F. & Lacy, L. Manipulating the Mouse Embryo: A Laboratory Manual 2nd edn (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1994)
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003)
Shiu, P. K., Raju, N. B., Zickler, D. & Metzenberg, R. L. Meiotic silencing by unpaired DNA. Cell 107, 905–916 (2001)
Nocka, K. et al. Molecular bases of dominant negative and loss of function mutations at the murine c-kit/white spotting locus: W37, Wv, W41 and W. EMBO J. 9, 1805–1813 (1990)
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998)
Lolle, S. J., Victor, J. L., Young, J. M. & Pruitt, R. E. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434, 505–509 (2005)
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001)
Sage, J. et al. Temporal and spatial control of the Sycp1 gene transcription in the mouse meiosis: regulatory elements active in the male are not sufficient for expression in the female gonad. Mech. Dev. 80, 29–39 (1999)
Acknowledgements
We are indebted to K. Thyagarajan for her participation in experimental work, J. J. Panthier for the gift of Kittm1Alf/+ mice and for helpful discussions, and K. B. Marcu and A. Schedl for help in preparing the manuscript. We thank M. Aupetit, Y. Fantei-Caujolle, J. P. Laugier, S. Pagnotta and K. Rassoulzadegan for expert technical assistance. This work was made possible by a grant to M.R. as ‘Equipe Labellisée’ of the ‘Ligue Nationale Française Contre le Cancer’.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Paternal and maternal transmission of paramutated alleles in 129/Sv and C57BL/6 genetic backgrounds. (DOC 39 kb)
Supplementary Table 2
Inheritance of the white-spotted phenotype induced by RNA microinjection. (DOC 42 kb)
Supplementary Figure 1
Inheritance of the white tail phenotype in crosses between Kit tm1Alf / + heterozygotes and paramutated homozygotes. (PDF 149 kb)
Supplementary Figure 2
Altered regulation of Kit transcription in the male germ cells of Kit tm1Alf / + heterozygotes. (PDF 48 kb)
Supplementary Figure Legends
Text to accompany the Supplementary Figures (DOC 29 kb)
Supplementary Methods
Oligonucleotide primers and miR sequences. Transcription run-on assay. (DOC 40 kb)
Rights and permissions
About this article
Cite this article
Rassoulzadegan, M., Grandjean, V., Gounon, P. et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474 (2006). https://doi.org/10.1038/nature04674
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04674
This article is cited by
-
Evidence base for non-genetic inheritance of environmental exposures in non-human animals and plants: a map of evidence syntheses with bibliometric analysis
Environmental Evidence (2023)
-
Missing Causality and Heritability of Autoimmune Hepatitis
Digestive Diseases and Sciences (2023)
-
Regulation of plant epigenetic memory in response to cold and heat stress: towards climate resilient agriculture
Functional & Integrative Genomics (2023)
-
Abundant small RNAs in the reproductive tissues and eggs of the honey bee, Apis mellifera
BMC Genomics (2022)
-
Small RNAs mediate transgenerational inheritance of genome-wide trans-acting epialleles in maize
Genome Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.