Scattered groups of these ancient fish may all stem from a single remote population.
Abstract
Coelacanths were discovered in the Comoros archipelago to the northwest of Madagascar in 1952. Since then, these rare, ancient fish have been found to the south off Mozambique, Madagascar and South Africa, and to the north off Kenya and Tanzania — but it was unclear whether these are separate populations or even subspecies. Here we show that the genetic variation between individuals from these different locations is unexpectedly low. Combined with earlier results from submersible and oceanographic observations1,2, our findings indicate that a separate African metapopulation is unlikely to have existed and that locations distant from the Comoros were probably inhabited relatively recently by either dead-end drifters or founders that originated in the Comoros.
Similar content being viewed by others
Main
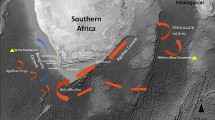
To estimate genetic variation, we analysed mitochondrial DNA sequences and microsatellite DNA from a total of 47 coelacanths (Latimeria chalumnae) from all of the different African locations apart from Tanzania (Fig. 1, red circles). (For methods, see supplementary information.) No differences were found in the mitochondrial sequences encoding cytochrome b, but six different haplotypes could be defined by nucleotide variations at various positions in the control region (for details, see supplementary information).
The three haplotypes not found in fish from the Comoros differ by only a single nucleotide from the most frequent Comoran haplotype, from which we infer that fish from all locations tested are closely related. (For comparison, we found a difference of 7.0–7.3% in the mitochondrial control-region nucleotide sequences of L. chalumnae and the Indonesian coelacanth L. menadoensis3.)
We also typed ten informative microsatellite loci using a total of 38 alleles (see supplementary information). No allele was restricted to fish from any one of the locations outside the Comoros. The relatedness between animals from the Comoros also extended to those from Mozambique, Madagascar, Kenya and South Africa. The genetic homogeneity indicated by this microsatellite analysis agrees with results from multilocus fingerprinting of samples from Mozambique and the Comoros4. Individual heterozygosity was low, which is generally taken as a sign of inbreeding5.
There were therefore no significant genetic differences in any of the coelacanths tested. Our results indicate that either all the specimens belong to a single, large, interbreeding (panmictic) population, or that there was a recent subdivision that could not be detected in our analysis. The shared mitochondrial haplotypes and microsatellite alleles in coelacanths from the Comoros and from other locations could be the result of free gene flow: however, the South Equatorial Current divides near the Comoros into the prevailing southerly Mozambique Current, which is dominated by a train of large anticyclonic eddies2, and into the northerly East African Coastal Current (Fig. 1), which might prevent gene flow from the south or the north to the Comoros.
Coelacanths remain at their home sites for many years1,6. On the basis of our observations from a submersible and our acoustic tracking experiments6, we believe that a regular benthic or pelagic migration across the deep ocean basins between the Comoros and the African or Madagascan mainland is highly unlikely, so a panmictic African population probably does not exist.
All coelacanths living outside the Comoros may have originated in the Comoros and/or other, as yet unknown localities in the Indian Ocean; they could be new arrivals, dead-end drifters (strays) or founders of young, reproducing subpopulations. Of 24 coelacanths we identified during surveys from our submersible off South Africa, two were gravid females, representative of a small founder population. And a female carrying developed eggs with visible embryos was also recorded among at least 19 coelacanths caught off Tanzania7.
It is possible that the western Indian Ocean could have been colonized relatively recently by coelacanth drifters from the Pacific province: this sluggish fish might be expected to survive long, passive oceanic transport8 by the Indonesian throughflow, which has probably existed for only about 3 million to 4 million years (ref. 9), into the South Equatorial Current that flows to the west. This idea is supported by molecular-clock evidence (our unpublished results) and by the low genetic diversity found in the Comoran and African populations.
References
Fricke, H. & Hissmann, K. Mar. Biol. 120, 171–180 (1994).
De Ruijter, W. P. M., Ridderinkhof, H., Lutjeharms, J. R. E., Schouten, M. W. & Veth, C. Geophys. Res. Lett. 29, 1502 (2002).
Holder, M. T., Erdmann, M. V., Wilcox, T. P., Caldwell, R. L. & Hillis, D. M. Proc. Natl Acad. Sci. USA 96, 12616–12620 (1999).
Schliewen, U., Fricke, H., Schartl, M., Epplen, J. T. & Pääbo, S. Nature 363, 405 (1993).
Keller, L. F. & Waller, D. M. Trends Ecol. Evol. 17, 230–241 (2002).
Hissmann, K., Fricke, H. & Schauer, J. Mar. Biol. 136, 943–952 (2000).
Durville, C. The Coelacanth, a ‘Fish Caught in Time’ http://www.jocara.net/Log_research/Coelacanth/coelacanth.html (2005).
Fricke, H. & Hissmann, K. Mar. Biol. 136, 379–386 (2000).
Cane, M. A. & Duncan, R. A. Nature 411, 157–162 (2001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Schartl, M., Hornung, U., Hissmann, K. et al. Relatedness among east African coelacanths. Nature 435, 901 (2005). https://doi.org/10.1038/435901a
Published:
Issue Date:
DOI: https://doi.org/10.1038/435901a
This article is cited by
-
A thirteen-million-year divergence between two lineages of Indonesian coelacanths
Scientific Reports (2020)
-
Single-male paternity in coelacanths
Nature Communications (2013)
-
The population biology of the living coelacanth studied over 21 years
Marine Biology (2011)
-
Mitochondrial genomic divergence in coelacanths (Latimeria): slow rate of evolution or recent speciation?
Marine Biology (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.