A search through the archives clears early vaccines of starting the AIDS pandemic.
Abstract
It has been suggested that chimpanzee kidney cultures may have been used in the preparation of oral polio vaccine stocks used in Africa during the late 1950s, and so could have introduced the primate precursor of the immunodeficiency virus HIV-1 into humans1,2. Here we analyse frozen samples of the suspect vaccine by using the polymerase chain reaction (PCR) to amplify any HIV-1-related nucleic acids or chimpanzee mitochondrial DNA that might be present, but we have failed to detect either. Our findings do not support the hypothesis that HIV-1 was introduced by oral vaccination against poliovirus.
Similar content being viewed by others
Main
A short description of five samples of oral polio vaccine (OPV) held by the Wistar Institute is given in Table 1. CHAT pool 13 was widely used to vaccinate thousands of people (mainly children) in Léopoldville3 in the former Belgian Congo (now Kinshasa, Democratic Republic of Congo) and some in Poland. Three further samples of CHAT vaccine held at the Centers for Disease Control (CDC) for 20–40 years were included in the study. The combinations of oligonucleotide primers and probes used can detect a wide variety of HIVs, including HIV-1 M, O and N isolates, as well as chimpanzee isolates Gab1, US and ANT70. The PCR assays have a sensitivity of about 50–80 RNA copies per ml. No sample proved positive for HIV-1-related nucleic acids by any of four primer pairs we used. By contrast, all samples were positive for poliovirus using PCR with reverse transcription, demonstrating that there was not extensive degradation of viral RNA (Table 1).
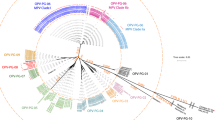
To identify the source of the kidney epithelial monolayers used to culture OPV, we analysed a small region (140 base pairs) within the mitochondrial 12S ribosomal RNA gene4. The primers we used amplify DNA from animals, including the African green monkey, chimpanzee, rhesus monkey and sooty mangabey (see supplementary information). All but one of the OPV samples were positive for mitochondrial (mt) DNA. With the exception of CHAT 1FL (see below), most sequences (86%) were highly related to those of the rhesus (Macaca mulatta) or cynomolgus monkey (Macaca fascicularis), and differed by only 0–2 bases from a known haplotype (Fig. 1).
The three groups correspond to the Macaca, Gorilla/Homo/Pan and Cercopithicus/Erythrocebus/Cercocebus lineages. Of the latter, C. atys and C. torquatus are Cercocebus lineage. Wistar and CDC OPV samples and mtDNA controls are in yellow, green and red, respectively; the Sabin-1 positive control is in blue. Other sequences are from databases. The phylogenetic tree was derived by the neighbour-joining method applied to pairwise sequence distances calculated using the Kimura two-parameter method transition/transversion ratio set to 10. Branch lengths are drawn to scale: bar, 0.1 nucleotide replacements per site. Numbers indicate the percentage of bootstrap samples in which the cluster is supported (only bootstrap values over 60% are given). Rhesus macaques are split into two groups, corresponding to the Chinese and Indian subspecies. Remaining sequences are nuclear insertions of mtDNA sequences (see supplementary information).
A few sequences were nuclear paralogues of mtDNA. The amplification of bona fide mtDNA sequences also picks up these nuclear insertions of mitochondrial sequences (NUMTs). They behave as pseudogenes and fix mutations more slowly than mtDNA itself5. A few of these NUMTs, three out of more than 250 sequences, were related to mtDNA sequences of a variety of Cercopithecus monkeys. These Cercopithecus-like sequences never exactly corresponded with a known sequence in the databases, but differed by about 4 base pairs. Most rhesus monkey mtDNA sequences corresponded to known mtDNA haplotypes. Some of the Cercopithecus-like sequences were also derived from single rhesus monkey samples (not shown). We conclude that these really are NUMTs and not evidence that some African monkey kidneys, along with those of macaques, had been used to grow OPV.
Given that OPV was prepared from pools of culture supernatants derived from individual pairs of kidneys, the presence of multiple mitochondrial haplotypes in the same sample is not unexpected. This would also explain the 3:2 mix of M. mulatta and M. fascicularis haplotypes in CHAT type 1 Wy4B. The detection of Homo sequences in CHAT 1FL stems from the fact that the virus was grown on the FL human diploid cell line.
To confirm these findings, we studied a mitochondrial 12S-rRNA locus (101 base pairs) just 5′ to the previous segment. This too was highly informative, with ten point substitutions and one insertion/deletion distinguishing macaque and chimpanzee sequences. All of our findings were confirmed. As the Wistar CHAT pool-13 lot was widely used in Léopoldville, we increased the resolution of the analysis. No chimpanzee sequences were found at a resolution of 1.7%. As this frequency is below the reciprocal of the number of animals generally used to make a lot of OPV, we conclude that none of these OPVs were prepared using chimpanzee kidneys.
In addition, none of the samples was positive when amplified with primers specific for chimpanzee nuclear DNA (data not shown). The extensive use of rhesus and cynomolgus monkeys in the preparation of all the Wistar samples described here not only confirms earlier claims6,7,8, as well as those denying the use of chimpanzee kidneys in the preparation of OPV9, but also reflects the procedures used in the late 1950s by Salk and Sabin.
CHAT pool 13 was used in Léopoldville between August 1958 and April 1960. The earliest bona fide HIV-1 sequence was derived from a 1959 serum sample taken from an adult living in the suburbs of Léopoldville10. According to the OPV/AIDS hypothesis1,2, the CHAT pool-13 sample should contain chimpanzee DNA, but as only macaque sequences were found, this tight geographical and temporal association would seem to have been a coincidence.
There is a corollary to our conclusion that the Wistar OPV samples were prepared using macaque kidneys. Given that simian immunodeficiency viruses (SIVs) are confined to African primates, the kidneys could not have been positive for SIV. In other words, the absence of SIV nucleic acids in all samples do not represent false negatives. We find no evidence to support the OPV/AIDS hypothesis.
References
Curtis, T. in Rolling Stone 54–60 (1992).
Hooper, E. The River: A Journey to the Source of HIV and AIDS (Penguin, London, 1999).
Plotkin, S. A., Lebrun, A., Courtois, G. & Koprowski, H. Bull. World Health Org. 24, 785–792 (1961).
van der Kuyl, A. C., Kuiken, C. L., Dekker, J. T. & Goudsmit, J. J. Mol. Evol. 40, 173–180 (1995).
Saitou, N. & Ueda, S. Mol. Biol. Evol. 11, 504–512 (1994).
Koprowski, H. J. Am. Med. Assoc. 178, 1151–1155 (1961).
Plotkin, S. A. in Proc. Sixth Intl Congr. Microbiol. Standardisation 48–73 (Hoffman, Berlin, 1961).
Plotkin, S. A., Katz, M., Brown, R. E. & Pagano, J. S. Am. J. Dis. Child. 111, 27–30 (1966).
Plotkin, S. A. & Koprowski, H. Science 286, 2450 (1999).
Zhu, T. et al. Nature 391, 531–532 (1998).
Hull, R. N., Cherry, W. R. & Tritch, O. J. J. Exp. Med. 165, 903–917 (1962).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary information
The online version of this article (doi:10.1038/35074171) contains supplementary material, which is available to authorized users.
Supplementary information
[This information is also available at: ftp://ftp.pasteur.fr/pub/retromol/wistar01/]
Methods
For the Alameda laboratory, two regions within the gag and pol genes of HIV-1/SIV were targeted for amplification. For the gag region, 3 upstream and 2 downstream primers were used to amplify a 155 bp fragment. The primers are all minor variants of the SK145-SKCC1B primers1 used in the AMPLICOR HIV-1 MONITOR version 1.5 Test. Two probes, SK102 and GAG102_O, were used to detect HIV-1 and SIVcpz (primarily US and ANT; and to a lesser extent GAB1), respectively. For the pol region, two upstream primers (one specifically for SIVcpzANT) and one downstream primer were used to amplify a 274 bp fragment. A single probe was used to detect both HIV-1 and SIVcpz.
Two hundred microliters of each sample were processed using the AMPLICOR HIV-1 MONITOR extraction protocol2. The equivalent of ~45 µl of the original sample, was analyzed in duplicate with both primer systems. One of the replicates for gag was amplified in the presence of 100 copies of IC to monitor inhibition; none was noted Following amplification, the gag products were analyzed on microwell plates coded with SK102, GAG102_O and, where appropriate, with the IC probe. The pol products were analyzed on plates coated with CC37.
Upstream gag primers were: WH43 AGTGGGGGGACATCAAGCAGCCATGCAAA, WH45 AGTGGGGGGACACCAGGCAGCTATGCAAA and SK145W AGTAGGGGGACATCAAGGAGCCATGCA. Downstream gag primers were: WH50 GGTACTAGTAGTTCCTGCTATGTCACTTCC and SKCC1W TGCTGGTTGTTCCTGCTATATCACTTCC. gag probes were SK102 GAGACCATCAATGAGGAAGCTGCAGAATGGGAT, GAG102_O GAAGTAATCAATGAGGAAGCAGCAGATTGGGAT. Upstream pol primers were: CC36B TAAAATTAGCAGGAAGATGGCCAGTAA and CC36Bant TAAAATTAGCCAGCAGATGGCCAGTAA. Downstream pol primers were: CC26B CCCCCAATCCCCCCTTTTCTTTTAAAATTGTG while the pol probe was CC37 TTTGGAATTCCCTACAATCCACAAAGTCAAGGAGT.
For the Paris laboratory total nucleic acids were extracted using Master Pure extraction kit (Epicentre) from half of the samples in order to keep some material left (total volumes varied from 300 µl to 1 ml). Briefly, 2 µg of tRNA (Sigma) were added as carrier and each sample was incubated 15 min at 56°C in the presence of proteinase K (100 µg/ml) then the acid nucleic lysates were purified. cDNA synthesis was performed on half of the extracts using UC1, UC12, cpzLTR3' and 3'ARN12S primers. To avoid HIV/SIV/polio contamination, the complete extraction and PCR amplifications procedures were performed in a HIV/SIV/polio free building.
Hot start PCR was applied in all reactions. The buffer conditions were as follows: 2.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 200 mM of each dNTP, 100 mM of each primer, 2.5 units of Taq DNA polymerase (Perkin Elmer) in a final volume of 100 ml. Annealing, extension and denaturation conditions were: an initial denaturation at 95°C for 5 minutes, then 35 cycles at 95°C (30 sec.), 55°C (30 sec.), 72°C (30 sec.) followed by a final step at 72°C for 10 minutes. Poliovirus VP1 (UG1 and UC1) and 3D (outer UG16 and UC12, inner UG7 and UC8) primers have been described3. Primers for SIVcpz were: cpzLTR5’ CTCAATAAAGCTTGCCTTGAGTG and cpzLTR3’ CCTGTTCGGGCGCCACTGCTAGAGAT. For mitochondrial 12S rDNA, 5'ARN12S CTATATACCGCCATCTTCAGCRAAC and 3’ARN12S TCTTRRTTTACTRCTAAATCCWCCTT as well as 12Sa and 12So4. PCR products were purified, cloned and sequenced.
References
-
1
Michael, N. L. et al. J. Clin. Microbiol. 37, 2557–2563 (1999).
-
2
Mulder, J. et al. J. Clin. Microbiol. 32, 292–300 (1994).
-
3
Guillot, S. et al. J. Virol. 74, 8434–8443 (2000).
-
4
Poinar, H. N. et al. Science 281, 402–406 (1998).
NUMT sequences
Chat pool 13
-
Cl 2
GCTTAGCCCTAAACTGTAATAGTTACATTAACAAAACTATTCACCAGAATACTACAAGCAATAGCTTAAAACTCAAAGGACTTGGTGGTGCTTTATGTCCCTC
-
Cl 6
GCTTAGCTGTAAACTTAAATAATTTAACAAACAAAATTATTCACCAGAGTATTACGAGCAAGAGCTTAAAACTCAAAGGACATGGCCATGCTTTATACTCCTC
-
Cl 8
GCTTAGCCTTAAACCTCAATAGTTAAAACAACAAAACTACTCTCCAGAATACTACAAGCAACAGCTTAAAACTCAAAGGACTTGGCGGTGCTTCACATCCCTC
-
Cl 10
GCTTAGCCCTAAACTCTAGTAGTTACATAAACAAAACAATTCCTCAGAATACTACAAGCAACGGCTTAAAACTCAAAGGACTTGGCGGTGCTTTGTATCCCTC
-
Cl 11
GCTCAGCTCTAAACTCTAATAGTTATATTAACAAAACCACTCGCCAGAGTACTACAAGAAACAGCTGAAAACTCAAAGGACTTGGTGGTGCTTTATATCCCCC
-
Cl 21
ACTTAGCCCTAAACTCTAATAGTTACATTAACAAAACCACTCACCAGAGTACTACGAGCAACAGCTTAAAACTCAAAGGATTTGGTGGTGCTTTATATCCGTC
-
Cl 23
GCTTAGCCCTAAACCTCAATAGTTAAAACAACAAAACTACTCACCAGAATACTACAAGCAACAGCTTAAAACTCAACGGACTTGGCGGTGCTTCACATCCCCC
-
Cl 24
GCTTAGCCCTAAACTGTAATAGTTACATTAACAAAACCATTAACCAGAATACTACAAGCAATAGCTTACAACTCAAAGGACTTGGTGGTGCTTTATGTCCCTC
-
Cl 27
GCTTAGCCCTAAAACTCAATAGTTAAATAACAAAACTACTCGCCAGAATACTACAAGCAACAGCTTAAAACTCAAAGGACTTGACGGTGCTTTACTTCCCTC
-
Cl 28
GCTTGGCCTTAAACCTCAGTAGTTAAATAACAAAACTACTTGCCAGAATACTACAAGCTACAGGTTAAAACTCAAAGGACTTGACGGTGCTTTACATCCCCC
-
Cl 30
GCTTAGTCCTAAACTTCATTAGTTAAATTAACAAAACTACTCACCAGAATACTACAAGCAATAGCTTAAAACTCAAAGGACCTGGCGGTGCTTTATATCCCCC
-
Cl 32
GCTTAGCCCTAAACTCTAGTAGTTACATAAACAAAACAATTCCTCAGAATACTACAAGCAACAGCTCAAAACTCAAAGGACTTGGCGGTGCTTTGTATCCCTC
-
Cl 33
GCTTAGCCCTAAACCTCAATAGTTAGAACAACAAAACTACTCGCCAGAATACTACAAGCAACAGCTTGAAACTCAAAGGACTTGGCAGTGCTTCATATCCCTC
-
Cl 38
GCTTAGTTCTAAACCCAAATAGTTCAACCAACAAAACTCTTCATCAGAGTACTATAAGCAACAGCTTAAGACTCAAAAGACTTGGTGGTGCTTTATATCCCTC
-
Cl 44
GCTTGGCCCTAAACCTCAGTAATTAGATAACAAAACTACTCGCCAGAATACTACAAGCAACAGCTTGAAACTCAAAGGACTTGACGGTGCTTTACATCCCCC
-
Cl 46
GCTTGGCCCTAAACTTCAACAATTAAATTAACAAAACTGCTCGCCAGAACACTACAAGCACTAGCTTAAAACTCAGAGGACTTGGCGGTGCTTCACATCCCTC
-
Cl 51
GCTCAGCTCTAAACTCTAATAGTTAGATTAACAAAACCACACGCCAGAGTACTACAAGAAACAGCTGAAAACTCAAAGGACTTGGTGGTGCTTTATATCCCCC
-
Cl 64
GCTTAGCCCTAAACCTCAATAATTAAAATAACAAAACTACTCGCCAGAATACTACAAGCAACAGCTTGAAACTCAAAGGACTTGGCGGTGTTTCACATCCCTC
Chat pool 16-A15
-
Cl 25
GCTTGGCCCTAAACCTCAGTAATTAGATAACAAAACTACTCGCCGAATACTACAGGCAACAGTTGAAACTCAAAGTACTTGACGGTGCTTTACATCCCCC
Wch24 57C-40
-
Cl 03
GCTTAGTTCTAAACCCAAATAGTTCAACCAACAAAACTCTCCATCAGAGTACTATAAGCAACAGCTTAAGACTCAAAAGACTTGGTGGTGCTTTATATCCCC
Chat type I Wy4B
-
Cl 148
GCTTGGCCCTAAACCTCAGTAATTAGATAACAAAACTACTCGCCAGAATACTACAAGCAACAGCTTGAAACTCAGAGGACTTGACGGTGCTTTACATCCCC
-
Cl 140
GCTTAGCCCTAAAACTCAATAGTTAAATAACAAAACTACTCGCCAGAATACTACAAGCAACAGCTTAAAACTCAAAGGACTTGACGGTGCTTTACATCCCC
Rights and permissions
About this article
Cite this article
Blancou, P., Vartanian, JP., Christopherson, C. et al. Polio vaccine samples not linked to AIDS. Nature 410, 1045–1046 (2001). https://doi.org/10.1038/35074171
Issue Date:
DOI: https://doi.org/10.1038/35074171
This article is cited by
-
Contaminated polio vaccine theory refuted
Nature (2004)
-
Non è Vero, è Mal Trovato: Polio Vaccine is not the cause of HIV
Rendiconti Lincei (2004)
-
Polio vaccines exonerated
Nature (2001)
-
The River without a paddle
Nature (2001)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.