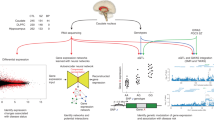
Abstract
Genome-wide association studies (GWAS) for schizophrenia have identified over 100 loci encoding >500 genes. It is unclear whether any of these genes, other than dopamine receptor D2, are immediately relevant to antipsychotic effects or represent novel antipsychotic targets. We applied an in vivo molecular approach to this question by performing RNA sequencing of brain tissue from mice chronically treated with the antipsychotic haloperidol or vehicle. We observed significant enrichments of haloperidol-regulated genes in schizophrenia GWAS loci and in schizophrenia-associated biological pathways. Our findings provide empirical support for overlap between genetic variation underlying the pathophysiology of schizophrenia and the molecular effects of a prototypical antipsychotic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Plenge RM, Scolnick EM, Altshuler D . Validating therapeutic targets through human genetics. Nat Rev Drug Discov 2013; 12: 581–594.
Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JE, Shah T et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet 2015; 385: 351–361.
Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005; 308: 385–389.
Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005; 308: 419–421.
Zareparsi S, Branham KE, Li M, Shah S, Klein RJ, Ott J et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet 2005; 77: 149–53.
Troutbeck R, Al-Qureshi S, Guymer RH . Therapeutic targeting of the complement system in age-related macular degeneration: a review. Clin Experiment Ophthalmol 2012; 40: 18–26.
McGrath J, Saha S, Chant D, Welham J . Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev 2008; 30: 67–76.
Mathers C, Fat DM, Boerma JT . The Global Burden of Disease: 2004 Update. Geneva, Switzerland: World Health Organization, 2008.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427.
Crowley J, Kim Y, Szatkiewicz J, Pratt A, Quackenbush C, Adkins D et al. Genome-wide association mapping of loci for antipsychotic-induced extrapyramidal symptoms in mice. Mammal Genome 2011; 23: 322–335.
Crowley JJ, Adkins D, Pratt A, Quackenbush C, van den Oord EJCG, Moy SS et al. Antipsychotic-induced vacuous chewing movements and extrapyramidal side-effects are highly heritable in mice. Pharmacogenomics J 2012; 12: 147–155.
Crowley JJ, Kim Y, Lenarcic AB, Quackenbush CR, Barrick CJ, Adkins DE et al. Genetics of adverse reactions to haloperidol in a mouse diallel: a drug-placebo experiment and Bayesian causal analysis. Genetics 2014; 196: 321–347.
Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010; 11: 733–739.
Creese I, Burt DR, Snyder SH . Dopamine receptor binding predicts clinical and pharmacological potencies of antischizophrenic drugs. Science 1976; 192: 481–483.
Crowley JJ, Zhabotynsky V, Sun W, Huang S, Pakatci IK, Kim Y et al. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat Genet 2015; 47: 353–360.
Fleischmann N, Christ G, Sclafani T, Melman A . The effect of ovariectomy and long-term estrogen replacement on bladder structure and function in the rat. J Urol 2002; 168: 1265–1268.
Turrone P, Remington G, Nobrega JN . The vacuous chewing movement (VCM) model of tardive dyskinesia revisited: is there a relationship to dopamine D(2) receptor occupancy? Neurosci Biobehav Rev 2002; 26: 361–380.
Keith F, George P . Paxinos and Franklin's The Mouse Brain in Stereotaxic Coordinates, 4th edn, Academic Press, 2013.
Trapnell C, Pachter L, Salzberg SL . TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25: 1105–1111.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Sun W, Liu Y, Crowley JJ, Chen TH, Zhou H, Chu H et al. IsoDOT detects differential RNA-isoform expression/usage with respect to a categorical or continuous covariate with high sensitivity and specificity. J Am Stat Assoc 2015; 110: 975–986.
Robinson MD, McCarthy DJ, Smyth GK . edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140.
Sun W . A statistical framework for eQTL mapping using RNA-seq data. Biometrics 2012; 68: 1–11.
Storey JD, Tibshirani R . Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003; 100: 9440–9445.
Sun W, Lee S, Zhabotynsky V, Zou F, Wright FA, Crowley JJ et al. Transcriptome atlases of mouse brain reveals differential expression across brain regions and genetic backgrounds. G3 2012; 2: 203–211.
Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011; 477: 289–294.
Leek JT, Storey JD . Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007; 3: 1724–1735.
Lee PH, O’Dushlaine C, Thomas B, Purcell S . InRich: interval-based enrichment analysis for genome-wide association studies. Bioinformatics 2012; 28: 1797–1799.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D . MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11: e1004219.
Genomes Project C Genomes Project C Auton A Genomes Project C Brooks LD Genomes Project C Durbin RM Genomes Project C Garrison EP Genomes Project C Kang HM et al. A global reference for human genetic variation. Nature 2015; 526: 68–74.
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983.
Major Depressive Disorder Working Group of the PGC. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013; 18: 497–511.
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013; 45: 1452–1458.
Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010; 467: 832–838.
Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012; 44: 981–990.
Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R . ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res 2011; 39 (Database issue): D712–D717.
Evsikov AV, Dolan ME, Genrich MP, Patek E, Bult CJ . MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol 2009; 10: R84.
Langfelder P, Horvath S . WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559.
Bernard V, Le Moine C, Bloch B . Striatal neurons express increased level of dopamine D2 receptor mRNA in response to haloperidol treatment: a quantitativein situhybridization study. Neuroscience 1991; 45: 117–126.
Kinkead B, Shahid S, Owens MJ, Nemeroff CB . Effects of acute and subchronic administration of typical and atypical antipsychotic drugs on the neurotensin system of the rat brain. J Pharmacol Exp Ther 2000; 295: 67–73.
Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O'Donovan MC et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry 2015; 20: 555–562.
Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 2008; 40: 827–834.
Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC et al. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep 2016; 16: 1126–1137.
Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015; 347: 1138–1142.
Gaspar HA, Breen G . Pathways analyses of schizophrenia GWAS focusing on known and novel drug targets. bioRxiv 2017; doi: https://doi.org/10.1101/091264.
Skene NG, Bryois J, Badden TE, Breen G, Crowley JJ, Gaspar HA et al Brain cell types and the genetic basis of schizophrenia (submitted).
Ase AR, Amdiss F, Hebert C, Huang N, van Gelder NM, Reader TA . Effects of antipsychotic drugs on dopamine and serotonin contents and metabolites, dopamine and serotonin transporters, and serotonin1A receptors. J Neural Transm 1999; 106: 75–105.
Meltzer HY, Li Z, Kaneda Y, Ichikawa J . Serotonin receptors: their key role in drugs to treat schizophrenia. Progr Neuro-psychopharmacol Biol Psychiatry 2003; 27: 1159–1172.
Faure P, Tolu S, Valverde S, Naude J . Role of nicotinic acetylcholine receptors in regulating dopamine neuron activity. Neuroscience 2014; 282C: 86–100.
Exley R, Clements MA, Hartung H, McIntosh JM, Cragg SJ . Alpha6-containing nicotinic acetylcholine receptors dominate the nicotine control of dopamine neurotransmission in nucleus accumbens. Neuropsychopharmacology 2008; 33: 2158–2166.
Drenan RM, Grady SR, Whiteaker P, McClure-Begley T, McKinney S, Miwa JM et al. In vivo activation of midbrain dopamine neurons via sensitized, high-affinity alpha 6 nicotinic acetylcholine receptors. Neuron 2008; 60: 123–136.
Lane RF, Blaha CD . Chronic haloperidol decreases dopamine release in striatum and nucleus accumbens in vivo: depolarization block as a possible mechanism of action. Brain Res Bull 1987; 18: 135–138.
Freedman R . alpha7-nicotinic acetylcholine receptor agonists for cognitive enhancement in schizophrenia. Annu Rev Med 2014; 65: 245–261.
Acknowledgements
This work was partially funded by the NIMH/NHGRI Center of Excellence for Genome Sciences grant (P50MH090338, P50HG006582, PIs Dr Fernando Pardo-Manuel de Villena and Dr Patrick F Sullivan).
Author contributions
PFS, YK, PG-R, FP-MdV and JJC designed the experiments. RJN, PG-R, AKR and CRQ performed the experiments. PFS, YK, PG-R, JJC, MDI-U, FP-MdV and PHL analyzed the data. JJC, PFS, YK and PG-R wrote the manuscript. All of the authors critically read and contributed comments to the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
PFS is a consultant to Pfizer. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Kim, Y., Giusti-Rodriguez, P., Crowley, J. et al. Comparative genomic evidence for the involvement of schizophrenia risk genes in antipsychotic effects. Mol Psychiatry 23, 708–712 (2018). https://doi.org/10.1038/mp.2017.111
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2017.111
This article is cited by
-
Transplantation of gut microbiota derived from patients with schizophrenia induces schizophrenia-like behaviors and dysregulated brain transcript response in mice
Schizophrenia (2024)
-
Molecular phenotypes associated with antipsychotic drugs in the human caudate nucleus
Molecular Psychiatry (2022)
-
Gene expression changes following chronic antipsychotic exposure in single cells from mouse striatum
Molecular Psychiatry (2022)
-
MiR-574-5P, miR-1827, and miR-4429 as Potential Biomarkers for Schizophrenia
Journal of Molecular Neuroscience (2022)
-
Analysis of the caudate nucleus transcriptome in individuals with schizophrenia highlights effects of antipsychotics and new risk genes
Nature Neuroscience (2022)