Abstract
Middle East and North Africa (MENA) encompass very unique populations, with a rich history and encompasses characteristic ethnic, linguistic and genetic diversity. The genetic diversity of MENA region has been largely unknown. The recent availability of whole-exome and whole-genome sequences from the region has made it possible to collect population-specific allele frequencies. The integration of data sets from this region would provide insights into the landscape of genetic variants in this region. We integrated genetic variants from multiple data sets systematically, available from this region to create a compendium of over 26 million genetic variations. The variants were systematically annotated and their allele frequencies in the data sets were computed and available as a web interface which enables quick query. As a proof of principle for application of the compendium for genetic epidemiology, we analyzed the allele frequencies for variants in transglutaminase 1 (TGM1) gene, associated with autosomal recessive lamellar ichthyosis. Our analysis revealed that the carrier frequency of selected variants differed widely with significant interethnic differences. To the best of our knowledge, al mena is the first and most comprehensive repertoire of genetic variations from the Arab, Middle Eastern and North African region. We hope al mena would accelerate Precision Medicine in the region.
Similar content being viewed by others
Introduction
The populations in the Middle East and North Africa (MENA) encompass over 7% of the world population1 and encompass significant ethnic, cultural, linguistic and genetic diversity. The populations in this region have in the past extensively admixed with populations of Asian, European, African continents, which have resulted in their rich diversity.2 An analysis of single-nucleotide polymorphism markers of over 270 individuals from Kuwait suggested extensive admixture with populations from Africa, Europe and Asia.3 The region has also been historically the melting pot for human migrations and modern civilization.4 A recent study using whole-exome sequences suggested that indigenous Arabs have been the first common ancestors of modern Eurasians, resulting from migration out of Africa.5
The region is characterized by a high prevalence of genetic diseases, contributed and aggravated by consanguinity. It is estimated that 25 to 60% of all marriages are consanguineous in the Arab world.6 A number of genetic diseases occurs specific to this part of the world including Familial Mediterranean Fever, which derives its name from the region.7 Several diseases and causative genes were also characterized for the first time from this part of the world.6 It has been widely believed that the high level of endogamy in the region would make the population ideal to study the genetic association, pathogenesis and prognosis of a number of genetic diseases8 underscoring the value in the genetic landscape of the population and its immense utility in medical genetics. A report by Tadmouri et al.9 suggest that certain dominant diseases are common to this population than elsewhere in the world. Systematic efforts to curate medically relevant genetic variants have also been underway through coordinated efforts. Recently, a well-structured catalog of genetic diseases has been made.10
One of the first personal genomes from the region was published from Kuwait, which included whole genomes and exomes of Bedouin ancestry.11 This was later followed up with whole genomes of an individual of Persian ancestry12 by the same group. Although a number of genome projects have been underway from MENA region, until very recently the genomic information was not publicly available, which limited their utility in clinical as well as epidemiological analysis.13 For example, the availability of whole-exome sequences from Qatar14 enabled us to analyze the landscape of pharmacogenetic variants for two common antithrombotic drugs—warfarin and clopidogrel.15 Similarly, large genome projects from the MENA region encompassing the Greater Middle East (GME) have provided a deep insight into the population structure and genetic variability of populations in the region.16
It has been previously suggested that the availability of a comprehensive resource of genetic variants and allele frequencies in the populations would enable and accelerate translational genomics in the region.13 Such a resource would enable cross-comparison of genetic variants, and allele and genotype frequencies across the data sets.
In the present report, we describe a comprehensive resource of human genetic variation, integrating whole-genome and whole-exome data sets from the MENA region. We catalog over 26 million genetic variants from Arab populations with its sub-populations. The resource has immense applications in understanding the allelic frequencies, carrier rates for rare genetic diseases and genetic traits including pharmacogenetics, apart from prioritizing and discovering novel disease-associated variants. The resource is publicly available at URL http://clingen.igib.res.in/almena.
Materials and methods
Data sets
Data set of whole-genome sequences from Qatar
We have used Qatar genome data set, which consisted of genome sequence variants of 108 individuals. (n=67), Persian/South Asian (n=23) and African (n=18) sub-populations of Qatar.5
Data set of whole-exome sequences from Qatar
Qatar exome data set consisted exome sequence variants of 100 individuals. The sub-population-wise breakup of the samples included (n=42), Persian/South Asian samples (n=33) and African samples (n=25).14
Data set of 1005 whole exomes and whole genomes from Qatar
This data set consisted of genome-sequenced variants of 88 individuals and exome-sequenced variants of 917 individuals, respectively. These individuals are from European (n=5), South Asian (n=76), Bedouin (n=490), African Pygmy (n=1), Arab (n=193), Persian (n=170) and sub-Saharan African (n=70).17
GME data set of whole-exome sequences
This data set encompassed a total of 1002 whole-exome sequences derived from multiple populations in the Middle East and North Africa. The samples were derived from individuals of Northwest Africa (n=99), Northeast Africa (n=368), Asian Peninsula (n=171), Israel (n=10), Syrian Desert (n=58), Turkish (n=164) and Central Asian (n=132) descent.16
Allele frequencies of Persians from Iran
This data set comprised of allele frequencies over six million variants found in 77 Iranian individuals. It was downloaded from https://irangenes.com/data-2/.
Creation of a unique compendium of genetic variants
All the data sets belonged to the assembly human genome 19 (GRCh37/hg19). The unique variants were retrieved from individual data sets and compiled into a compendium.
Annotation of the variants
The unique sets of 26 million variants were systematically annotated across a number of public databases and computational algorithms using ANNOVAR.18 A total of 33 tools as detailed in Supplementary Data 3 were screened for the variants using ANNOVAR Perl scripts.18 The summary of the data integration and annotation pipeline is detailed in Figure 1.
Allele and genotype frequencies
The variant call files were used to calculate allele and genotype frequencies with the open-source whole-genome association analysis toolset PLINK1.919 to compute the allele and genotype frequencies. If data sets were not available in variant call formats, the allele and genotype frequencies were directly retrieved from the supplementary files associated with the original publication or web resource.
Database and web server
The data was ported onto a scalable database system MongoDB, extensively used and a popular open-source NOSQL database system widely used for big data sets. The web interface was coded in Perl/CGI and Javascript. The web server was configured in Apache 2.4.12.
Demonstration on application of server with TGM1 variants
To demonstrate the application of database, we took TGM1 variants for performing possible analyses. A list of pathogenic variants in TGM1 was downloaded from ClinVar, a comprehensive online resource of clinically significant genetic variants. This list encompassed a total of 37 variants. Of these, a total of 34 variants were annotated as pathogenic (marked CLNSIG=5). The allele frequencies for these variants were compared in individual populations using the database. Test of significance was carried out for the allele frequency of the variants belonged to gene of interest using Fisher's test compared with the allele count of 1000 genome project from Ensembl browser (http://grch37.ensembl.org/index.html).
Results
Creation of a compendium of unique variants integrating data from multiple data sets
Integrating data sets, we assembled a unique compendium of 26 828 057 genetic variants, of which 19 210 183 were already known variants. The genome data sets contributed to a significant amount of the variants, with 23 354 688 variants contributed by the Qatar 108 whole-genome data set. Table 1 summarizes the data sets and genetic variants integrated in the compendium. It also briefs the corresponding sequencing methods, and the number of sub-populations included in each data set. A description on methods, sequencing platform and filtering options are included in Supplementary Data 1.
Annotation of the genetic variants
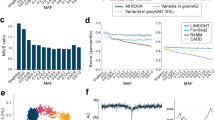
The genetic variants were systematically annotated by various databases and tools in ANNOVAR package such as dbSNP142, clinvar2016, refseq, 1000 genome (2015.Aug), esp6500, exac03, GWAS catalog and cytoband. Annotation of the data sets revealed a total of 11 493 833 mapped to genic regions, whereas 13 726 198 variants were mapped to intergenic regions in the genome. Of the variants in the genic boundaries, a total of 767 241 were exonic and 8 783 767 were intronic in origin. Supplementary Data 2 shows basic refgene annotation of variants. The overview of the variants and distribution is summarized in Figure 2.
Summary distribution of the variants in the al mena compendium of genetic variants. (a) Overlaps and contributions of variations from the three major data sets and (b) genomic context of the genetic variants in the database. A full color version of this figure is available at the Journal of Human Genetics journal online.
Our analysis revealed a total of 455 251 variants, which were nonsynonymous in nature. The mapping statistics of genetic variants of our compendium is given in Supplementary Data 1. The pathogenicity of the variants was annotated using ANNOVAR’S ljb_all database annotation. This data set includes the computational predictions of pathogenicity of variants such as SIFT,20 Polphen2,21 MutationTaster22 and MutationAssessor.23 Supplementary Data 3 provides description and interpretation of computational tools for predicting pathogenicity of the variant.
In addition, variants were systematically annotated across a number of relevant databases. These include ClinVar24 for clinically relevant variants implicated in Mendelian genetic diseases. A total of 1325 variants mapped to known pathogenic variants from ClinVar2016 database.
Web-based interface for query and analysis
Toward enabling quick access to the variants, allele and genotype frequencies and relevant annotations, a web-based interface to the resource was created. The interface was designed to be user-friendly. The search/query box can be used to query the resource using specific genetic variant IDs (rsIDs), gene names, positions or ranges of genomic positions. The interface returns a neatly organized list of links where the user can find more information on the specific variants that match the query condition. The variant page provides details about the variant, its genomic context, population frequency across different populations in the MENA region as well as across the 1000 genome data sets and populations. Functional and clinical annotations for each variant have also been precomputed and available. The user can see the description of each field to ease the understanding of algorithms and data sets used for annotation. The variants are also linked out to relevant databases including UCSC Genome Resource,25 dbSNP26 and ClinVar. Figure 3 summarizes the information available for a single variant in the resource.
Proof-of-principle application of the variant resource in genetic epidemiology
As a proof-of-principle application of the compendium of genetic variations, we evaluated the genetic epidemiology of variants in TGM1 gene, associated with autosomal recessive lamellar ichthyosis in the database. As detailed in the Materials and methods section, the list of single-nucleotide variants annotated ‘pathogenic’ for the gene TGM1 were downloaded and searched in the database. Out of a total of 37 variants in the ClinVar database for TGM1 gene, we retrieved a total of 34 variants, which were marked pathogenic. These variants were queried across the compendium. A total of four variants mapped to the compendium. The allele and genotype frequencies of the variants are listed in Table 2.
Our analysis suggests the allele frequency for pathogenic variants ranged from 0.001 to 0.018 across the data sets considered. On an average, this translates to a carrier rate of over 0.0095 in the populations considered.
Our analysis also reveals that all four pathogenic variants are significantly different in Northeast or Northwest African region and S42Y is found significant in Arabian Peninsula and Northeast Africa, compared with the 1000 genome allele frequencies. This suggests an assay of just four variants, which would enable cheap, cost-effective carrier screening, prenatal and neonatal screening and postnatal diagnosis in the region.
Discussion
The MENA region, especially the Mediterranean Basin has been the hotbed for human migration providing an interesting area to understand human population genetics.4 The recent availability of whole-genome and whole-exome data sets from the Mediterranean region has significantly improved our understanding of the admixture as well as the natural history of human migrations.5 In addition, it has also significantly improved our understanding of the genetic diversity and a hitherto uncharacterized repertoire of human genetic variations.
The understanding of human genetic variations from the MENA region is believed to have significant impact on the identification of new disease-associated variants16 with the clinical relevance in the population and sub-populations.15 The availability of population-level allele frequency data in public domain helps researchers to compare and analyze clinically relevant variations toward achieving quick translation.
In the present report, we have integrated whole-genome and whole-exome data sets for over 2000 individuals from 15 sub-populations to create a unique compendium of genetic variants from Arab population. Our analysis uncovered a total of over 26 million unique genetic variants, out of which over a six million genetic variants have not been previously represented in any major databases including dbSNP, 1000 genome or ExAC. Allele and genotype frequencies for the variants were computed across the populations, which enabled understanding the landscape of genetic variants.
The Greater Middle East (GME) Variome Project has made available a web-based server for exome variants from different regions generated as part of a multinational collaboration to generate a reference population for the GME.16 Our resource encompasses a number of features, annotations and data sets, which have not been part of this database. Supplementary Data 4 summarizes the differences and unique features of al mena compared with this resource. The al mena resource presently enable users to have a comparison of carrier frequency of mutant alleles across 15 sub-populations.
As a proof of concept for the utility of the resource in clinical and genetic epidemiological studies, we evaluated the allele frequencies of clinically relevant variants associated with autosomal recessive lamellar ichthyosis caused by genetic variations in TGM1 gene. The protein product of TGM1 gene, transglutaminase, catalyses the formation of ɛ-(γ-glutamyl)-lysine crosslinks in proteins and thereby stabilizes the biological structures. TGM1 mutations prevent the protein from forming the cornified cell envelope, thereby causing lamellar ichthyosis, a condition that causes extensive scaling of skin in addition to other skin abnormalities.27 Mutations either in a homozygous or compound heterozygous form in the TGM1 gene have been previously reported in Arab populations.28, 29 Our analysis suggests that the four TGM1 pathogenic variants, which mapped to the compendium, have significantly distinct allele frequencies in the sub-populations considered. In our study, this mutation has significantly stood out from 1000 genome allele frequency at 3.0e−3 P-value with Northeast African population and 1.0e−4 P-value in Arabian Peninsula-specific population. Our analysis also suggests a high frequency of the variants ranging from 1.6 in 100 to 18/1000.
This study highlights the need for systematic analysis of clinically relevant genetic variants in the populations and how the availability of a well-curated database would quickly enable the translational applications of genomic data toward benefitting to estimate disease burden, carrier rates and possible policies toward accurate, fast and cost-effective diagnosis.
A number of genome projects are presently underway in the region.13 We hope al mena would be enriched with larger data sets encompassing multiple sub-populations, which are not yet included in the database currently. To the best of our knowledge, al mena is a unique resource for genetic variants in Arab, Middle East and North Africa and a pioneering step toward enabling Precision Medicine in the region.
References
Parkash, J., Younis, M. Z. & Ward, W. Healthcare for the ageing populations of countries of Middle East and North Africa. Ageing Int. 40, 3–12 (2015).
Yang, X., Al-Bustan, S., Feng, Q., Guo, W., Ma, Z., Marafie, M. et al. The influence of admixture and consanguinity on population genetic diversity in Middle East. J. Hum. Genet. 59, 615–622 (2014).
Alsmadi, O., Thareja, G., Alkayal, F., Rajagopalan, R., John, S. E., Hebbar, P. et al. Genetic substructure of Kuwaiti population reveals migration history. PLoS ONE 8, e74913 (2013).
Poulain, M. Migratory flows in the Mediterranean Basi. Polit. Etrang. 59, 689–705 (1994).
Rodriguez-Flores, J. L., Fakhro, K., Agosto-Perez, F., Ramstetter, M. D., Arbiza, L., Vincent, T. L. et al. Indigenous Arabs are descendants of the earliest split from ancient Eurasian populations. Genome Res. 26, 151–162 (2016).
Al-Gazali, L., Hamamy, H. & Al-Arrayad, S. Genetic disorders in the Arab world. BMJ 333 (2006).
Belmahi, L., Sefiani, A., Fouveau, C., Feingold, J., Delpech, M., Grateau, G. et al. Prevalence and distribution of MEFV mutations among Arabs from the Maghreb patients suffering from familial Mediterranean fever. C. R. Biol. 329, 71–74 (2006).
Zayed, H. The Arab genome: health and wealth. Gene 592, 239–243 (2016).
Tadmouri, G. O., Nair, P., Obeid, T., Al Ali, M. T., Al Khaja, N. & Hamamy, H. A. Consanguinity and reproductive health among Arabs. Reprod. Health 6, 17 (2009).
Tadmouri, G. O. CTGA: the database for genetic disorders in Arab populations. Nucleic Acids Res. 34, D602–D606 (2006).
Alsmadi, O., John, S. E., Thareja, G., Hebbar, P., Antony, D., Behbehani, K. et al. Genome at juncture of early human migration: a systematic analysis of two whole genomes and thirteen exomes from Kuwaiti population subgroup of inferred Saudi Arabian Tribe Ancestry. PLoS ONE 9, e99069 (2014).
Thareja, G., John, S. E., Hebbar, P., Behbehani, K., Thanaraj, T. A. & Alsmadi, O. Sequence and analysis of a whole genome from Kuwaiti population subgroup of Persian ancestry. BMC Genomics 16, 92 (2015).
Al-Mulla, F. The locked genomes: A perspective from Arabia. Appl. Transl. Genomics 3, 132–133 (2014).
Rodriguez-Flores, J. L., Fakhro, K., Hackett, N. R., Salit, J., Fuller, J., Agosto-Perez, F. et al. Exome Sequencing identifies potential risk variants for Mendelian disorders at high prevalence in Qatar. Hum. Mutat. 35, 105–116 (2014).
Sivadas, A., Sharma, P. & Scaria, V. Landscape of warfarin and clopidogrel pharmacogenetic variants in Qatari population from whole exome datasets. Pharmacogenomics 17, 1891–1901 (2016).
Özçelik, T. & Onat, O. E. Genomic landscape of the Greater Middle East. Nat. Genet. 48, 978–979 (2016).
Fakhro, K. A., Staudt, M. R., Ramstetter, M. D., Robay, A., Malek, J. A., Badii, R. et al. The Qatar genome: a population-specific tool for precision medicine in the Middle East. Hum. Genome Var. 3, 16016 (2016).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164–e164 (2010).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet (Chapter 7, Unit 7.20) (2013).
Schwarz, J. M., Rödelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118–e118 (2011).
Landrum, M. J., Lee, J. M., Riley, G. R., Jang, W., Rubinstein, W. S., Church, D. M. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Karolchik, D., Hinrichs, A. S. & Kent, W. J. The UCSC Genome Browser. Curr. Protoc. Bioinform (Chapter 1, Unit 1.4) (2009).
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Eckert, R. L., Sturniolo, M. T., Broome, A.-M., Ruse, M. & Rorke, E. A. Transglutaminase function in epidermis. J. Invest. Dermatol. 124, 481–492 (2005).
Cserhalmi-Friedman, P. B., Milstone, L. M. & Christiano, A. M. Diagnosis of autosomal recessive lamellar ichthyosis with mutations in the TGM1 gene. Br. J. Dermatol. 144, 726–730 (2001).
Huber, M., Rettler, I., Bernasconi, K., Frenk, E., Lavrijsen, S. P., Ponec, M. et al. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science 267, 525–528 (1995).
Acknowledgements
We acknowledge the constructive criticism and suggestions from Dr Srinivasan Ramachandran and Judith Hariprakash. The work was funded by the Council of Scientific and Industrial Research (CSIR), India through Grant BSC0212 (Wellness Genomics Project).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Koshy, R., Ranawat, A. & Scaria, V. al mena: a comprehensive resource of human genetic variants integrating genomes and exomes from Arab, Middle Eastern and North African populations. J Hum Genet 62, 889–894 (2017). https://doi.org/10.1038/jhg.2017.67
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.67
This article is cited by
-
Ichthyosis
Nature Reviews Disease Primers (2023)
-
Health inequity in genomic personalized medicine in underrepresented populations: a look at the current evidence
Functional & Integrative Genomics (2023)
-
Genomic data integration and user-defined sample-set extraction for population variant analysis
BMC Bioinformatics (2022)
-
The diagnostic yield, candidate genes, and pitfalls for a genetic study of intellectual disability in 118 middle eastern families
Scientific Reports (2022)
-
Genomics of rare genetic diseases—experiences from India
Human Genomics (2019)