Abstract
Lantibiotics are biologically active peptides produced by Gram-positive bacteria. Starting from fermentation broth extracts preselected from a high-throughput screening program for discovering cell-wall inhibitors, we successfully isolated a new lantibiotic produced by Actinoplanes sp., designated as NAI-802. MS and NMR analysis together with explorative chemistry established that NAI-802 consists of 21 amino acids, 19 of which are identical to those present in the class II lantibiotic actagardine. Interestingly, NAI-802 carries one extra alanine and one extra arginine at the N- and C-termini, respectively. As expected from the overall higher positive charge, NAI-802 was slightly more active than actagardine against staphylococci and streptococci. Further improvement of its antibacterial activity was achieved by adding one additional positive charge through conversion of the C-terminal carboxylate into the corresponding basic amide. NAI-802 thus represents a novel promising candidate for treating Gram-positive infections caused by multidrug-resistant pathogens.
Similar content being viewed by others
Introduction
The lantibiotic NAI-107 (previously known as 107891 or microbisporicin), produced by Microbispora sp.,1 displays potent activity against most Gram-positive pathogens of medical importance2 and is effective in experimental models of infection.3 This compound was isolated after high-throughput screening (HTS) of ca. 120 000 microbial broth extracts through a program for identification of compounds active on bacterial cell wall. Unexpectedly, this assay system turned out to significantly enrich for lantibiotics.4 We thus decided to further investigate the samples positive in that screening program and report here the isolation, structure elucidation and preliminary biological profiling of a new lantibiotic, designated NAI-802.
Our proprietary Natural Products Library database contains information regarding strains, their growth conditions (media, growth temperatures and harvest times) and extracts (preparation procedure). It also reports data obtained with extracts in the different HTS programs run at Biosearch Italia/Vicuron Pharmaceuticals in the 1996–2006 period. Overall, it covers about 70 000 strains, 160 000 extracts and over 20 HTS programs. Details of the screening project for identification of compounds active on bacterial cell wall have been described elsewhere.4 Briefly, the program involved the HTS microbial extracts to identify those able to inhibit Staphylococcus aureus growth but ineffective against its isogenic L-forms (protoplast-type cells that are able to replicate under appropriate osmotic conditions despite the lack of a functional cell wall and are thus insensitive to peptidoglycan synthesis inhibitors). Only those extracts were further retained whose activity against S. aureus was not affected by incubation with β-lactamases or with excess ɛ-amino-caproyl-D-alanyl-D-alanine. The extracts generated from the actinomycete strains IDs 104802 and 104771 were among those fulfilling the screening selection criteria.
Results
Taxonomy of the producing strains
We report here on the analysis of the actinomycete strains reported as ID104802 and ID104771, and classified as Actinoplanes on the basis of morphological features and formation of motile spores. Actinoplanes strains ID104802 and ID104771 were deposited as Actinoplanes sp. DSM24057 and DSM25201, respectively, in Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). When grown on S1 medium, both strains formed dark-orange colonies with whitish aerial mycelium. A brown–green pigment was released in the medium upon ageing of the cultures. Nearly complete sequences (1439 bp) of the 16 S ribosomal RNA gene showed that the two strains are closely related having almost identical sequences (>99.9%). Comparison of the sequences with those maintained in GenBank showed >98% identity with the 16 S ribosomal RNA gene sequences of several Actinoplanes species (highest identity with A. teichomyceticus), thus confirming the original classification. Accession numbers for the 16 S sequences of strains ID104771 and ID104802 are JX680596 and JX680597, respectively.
Fermentation, isolation and purification of NAI-802
For metabolite analysis, the strains were grown in M8 medium for 168 h and the corresponding cultures assayed for antimicrobial activity. After HPLC fractionations of the broth filtrates, both strains yielded a single fraction endowed with activity against Bacillus subtilis 168, eluting at 17.6 min. Upon HR-MS, the 17.6-min fraction from both strains showed a double protonated ion at m/z 1058.9707, together with a triple protonated ion at m/z 706.3151, thus corresponding to a molecular mass of 2116 a.m.u. and suggesting a molecular formula of C90H141N25O26S4. No matches were found between this mass and known molecules reported in the proprietary ABL database, which contains UV properties and mass data on ∼29 000 microbial metabolites published up to 2006.5 Further experiments using the production media M8 and KC4 indicated that the 2116 a.m.u. compound was detectable after about 48 h, reaching maximum production after 72–96 or 96–120 h in media KC4 and M8, respectively, for both strains. Production by both strains in medium KC4 was ca. three times higher than in M8, as estimated from the relative area of the HPLC peaks. After growing strain ID104802 in KC4 medium for 72 h in a 20-l fermentor, the culture was harvested and the mycelium was separated from the supernatant broth by filtration. HPLC analysis showed that the mass 2116 peak was equally distributed between the broth filtrate and the mycelium, and so was the activity against B. subtilis 168. The work-up procedure involved methanol extraction of the mycelium and HP20 resin adsorption of the broth filtrate, followed by reversed-phase chromatography, as described under Experimental Procedure. This procedure allowed the recovery and purification of pure NAI-802 1. During the recovery process, we also observed minor amounts of a related congener designated Ala(0)-NAI-802 2, as described below.
Structure determination and physico-chemical properties
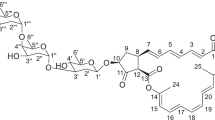
The structure of 1 (Figure 1) was elucidated by spectroscopic analysis and chemical studies. Its UV spectrum exhibits two shoulders at 225 and 280 nm, as usually found in peptides, while its 1H NMR spectrum displayed a typical peptide pattern (Supplementary Figure 1). The correlation of 1H NMR signals to the corresponding 13C-carbon atoms was carried out in a HSQC NMR experiment, followed by analysis of the COSY and HMBC spectra. These data indicated the absence of dehydrated residues such as dehydroalanine or dehydrobutyrine, while the presence of a spin system attributed to one Trp residue could be easily established by NOE and HMBC contacts from the β-methylene group to the indole ring. The presence of one Leu, two Val and two Ile residues could be readily assigned by their characteristic 1H and 13C chemical shifts, while the presence of one Ser was assigned by the characteristic 13C of the β-methylene group in the HSQC spectrum. One Arg was clearly identified from its particular spin system. As observed for actagardine,6 the HSQC spectra revealed unusual 13C chemical shifts of the β- and γ-carbons of one of the β-methyl-lanthionine (Me-Lan) bridges, suggesting the presence of one oxidized Me-Lan bridge in 1. To partially assign the peptide sequence, we investigated short-range NOESY cross peaks between the CHα, CHβ or amide proton of residue i and the amide proton of the adjacent i+1 residue in the peptide sequence, as described.7 Accordingly, we were able to sequentially assign part of the spin systems establishing an overall sequence identity with actagardine while the C-terminal position of the Arg residue in 1 was highlighted by the NOESY correlation between Arg-NH at 8.29 and Lan-20 CHα at 5.14. The assignments of protons and carbons in the NMR spectra of NAI-802 are summarized in Table 1.
MS analyses were performed on the hydrolyzed and intact compound. Analysis of the acid hydrolysate showed the presence of Lan(1), Me-Lan (3), Ala (2), Arg (1), Gly (2), Ser (1), Trp (1), Val (2), Glu (1), Leu (1) and Ile (2), consistent with NMR analyses. The HR-mass spectrum of intact 1 showed the presence of two intense signals at m/z 1058.9707 and 706.3151 corresponding to the double- [M+2H]2+ and triple- [M+3H]3+ charged ions, respectively. Upon MS/MS fragmentation of the m/z 1059 [M+2H]2+ ion at several collision energy values, the double protonated ion at m/z 1050, resulting from a single dehydration, was initially formed, followed by two major monoprotonated ions at m/z 673 and 1445, consistent with residues 1–7 and 8–21 of the assigned structure (Table 2). Ions at m/z 870 and 1248, consistent with residues 1–9 and 10–21, were also formed, together with ions m/z 1856 and 1786, consistent with dehydrated residues 1–19 and 1–18 (or 2–19), respectively (Table 2), thus confirming the C-terminal position of the Arg residue. The structure established by spectroscopic means was confirmed by selected chemical analyses. Edman degradation of 1 established Ala as the N-terminal residue. Reaction with ethanethiol at neutral pH excluded the presence of dehydrobutyrine, dehydroalanine or disulfide bridges,8 whereas the same reaction at alkaline pH suggested the presence of four Lan bridges from the addition of four mercaptoethanol moieties to 19 after reduction of the sulfoxide, which in turn is expected to occur under alkaline conditions as previously reported for actagardine.6 Moreover, two carboxylic acids, associated with the single Glu and the C-terminal Arg residues, were detected following amidation of 1 with benzylamine and isolation of the resulting diamide derivative. However, when ethylendiamine replaced benzylamine, the amidation reaction occurred regioselectively at the C-terminus, yielding the monoamidated derivative 3, as indicated by the observation of two intense signals at m/z 1080 and 720, corresponding to the double [M+2H]2+ and triple [M+3H]3+ charged ions, respectively, of 3 (Figure 2 Supplementary data) and by the fragmentation pattern (Table 2). Careful inspection by LC-MS of partially purified samples of 1 highlighted the presence of an impurity coeluting with 1 at 17.6 min. Its HR-mass (m/z 1094.4889 [M+2H]2+) was consistent with the formula C93H146N26O27S4 and the presence of an additional Ala residue, while the MS/MS fragmentation yielded major peaks at m/z 745-1445 and 942-1248 (Table 2). These results suggested that this impurity is actually a minor congener of 1 carrying an extra Ala at the N-terminus, and was thus designated Ala(0)-NAI-802 2. Both Actinoplanes sp. ID104802 and Actinoplanes sp. ID104771 produced 1 and 2 in an approximate ratio of 20:1. An Ala(0)-congener has also been reported for actagardine.10
Biological properties
The in vitro activities of 1 and 3 against a panel of microbial strains is reported in Table 3. 1 was active against staphylococci and streptococci, with MICs ranging from 0.5 to 32 μg ml−1, while the MICs against Enterococcus sp. were 128 μg ml−1 or higher. The compound was not active against Escherichia coli or Candida albicans. Overall, 1 showed a better activity than actagardine, with MIC values consistently 2–4 fold lower against Staphylococcus and Streptococcus sp. Compound 3 exhibited an improved antimicrobial activity, with MIC values reduced 4–8 fold in comparison to 1, leading also to measurable MICs against enterococci. When measurable, the MICs of 1, 3 and actagardine were not significantly affected by the presence of 30% bovine serum in the media or by the resistant phenotypes of the strains. Compounds 1 and 3 showed a significant activity also against Gram-positive anaerobic bacteria, such as Clostridium difficile, C. butyricum, C. perfringens and Peptostreptococcus asaccharolyticus with MIC ranges of 0.25–2 and ⩽8 μg ml−1 for 1 and 3, respectively. These MIC values are comparable to vancomycin’s (Table 4). As expected, no activity was observed against the Gram-negative anaerobe Bacteroides fragilis. It should be noted that, while 3 was 2–4 times more active than 1 against the three C. difficile strains, it was 2–4 less active against the three other tested Clostridium spp. (Table 4).
Discussion
In the last two decades the ribosomally synthesized lantibiotics have aroused interest for their antibacterial activity, particularly against the worrisome methicillin-resistant S. aureus.11, 12 Approximately 60 lantibiotics, mostly produced by strains belonging to the phylum Firmicutes, have been discovered since 1927 when the first representative, nisin, was discovered,13 but relatively few from the Actinobacteria, possibly because of the abundance of many other classes of lower MW antibiotics produced by this phylum. Recent investigations are, however, demonstrating that lantipeptides (Lan-containing peptides called lantibiotics when they posses antibiotic activity) can be frequently found in other bacterial phyla, including the Actinobacteria, expanding the chemical diversity and the biological activities of this family of ribosomally synthesized peptides.14 In the present study, we show that two Actinoplanes strains, ID104802 and ID104771, produce the new lantibiotic NAI-802 1, along with minor amounts of its Ala(0)-congener 2. Compound 1 is highly related to the class II lantibiotic actagardine and thus represents a further addition in this family (Figure 2), which includes: actagardine and Ala(0)-actagardine, produced by Actinoplanes liguriae ATCC 31048;15 actagardine B, produced by A. liguriae NCIMB41362;16 michiganin A, produced by Clavibacter michiganensis;17 NAI-802 and Ala(0)-NAI-802, as described here. All these lantibiotics share three conserved thioether bridges within a core 19-aa peptide consisting of 14 invariant residues. Actually, the number of invariant residues would be 16 if those present in the precursor peptide were taken into account, as michiganin lacks one of the central Lan bridges despite possessing the amino-acid residues (dehydrobutyrine and Cys) required for its formation (Figure 2). Among the natural variants described so far, variable positions accommodate hydrophobic residues only. However, many positions can be manipulated by appropriately engineering the structural gene encoding the precursor peptide of actagardine in the producing strain.16 Despite the high conservation of the central core in the natural variants, there appears to be some flexibility at the N- and C-terminal ends of this lantibiotic family. This feature, which had been adumbrated by the discovery of Ala(0)-actagardine,10 became apparent with the discovery of michiganin A, which carries extra Ser and Arg residues at the N- and C-termini, respectively,17, 18 and has further been confirmed in this study with 1, where Ala and Arg are found, respectively, and its Ala(0)-congener 2. It remains to be determined whether N- and/or C-terminally extended variants could be easily generated by appropriately engineering the actagardine structural gene. Considering its high structural similarity to actagardine, 1 is likely to exert its antimicrobial action through a similar mechanism by binding to the peptidoglycan intermediate lipid II, as does actagardine.19 A conserved Glu residue (Figure 2) is present in many class II lantibiotics (such as mersacidin, lacticin 481, lacticin 3147 A1 and haloduracin-α) and has been demonstrated to be essential for antibiotic activity in mersacidin.20 In comparison with actagardine, 1 carries one additional positive charge at physiological pH due to the C-terminal Arg, a feature that might explain its improved antimicrobial activity (Table 3). Positively charged residues have indeed been suggested to have an important role in the interaction of lantibiotics with the negatively charged bacterial membranes.20 The antibacterial activity of actagardine was improved after transforming the negatively charged C-terminal carboxylate into a positively charged basic amide.21 The same result is observed here with the basic amide derivative 3, indicating an additive effect on antimicrobial activity of the basic amide and of the Arg side chain. Compound 3 resulted particularly active against Streptococcus sp. It is worth recalling that, despite the presence of two carboxylates, only the C-terminus reacted with a diamine, giving the corresponding monoamide. An identical behavior was observed with actagardine under similar conditions.21 Apparently, di-carboxamide derivatives of actagardine and 1 can be obtained only with a monoamine but not with a diamine, even under a stechiometric excess of reactants (data not shown). Those outcomes suggest that a different conformation of NAI-802, depending on the reactive amine, allows or prevents reaction on the glutamic residue. Yet deeper studies should be necessary to clarify the reason of this behavior.
In conclusion, NAI-802 and its derivative 3 obtained by regio-selective amidation represent promising class II lantibiotics for treating infections caused by multidrug-resistan Gram-positive pathogens. Indeed, a related compound NVB302, developed for treating C. difficile-associate diarrhaea, is undergoing formal preclinical studies.22
Experimental procedure
General experimental conditions
HPLC chromatography employed a Shimadzu Series 10 spectrophotometer (Shimadzu Corporation, Kyoto, Japan), equipped with a Lichrosphere C18 4.6 × 100 mm column (Merck, Darmstadt, Germany) and a diode-array detector (190–800 nm), using a linear gradient of 0.05 M ammonium formate-acetonitrile (from 5 to 90% of organic phase in 30 min) at a flow rate of 1 ml min−1. ESI-MS data were recorded on an Ion Trap ESQUIRE 3000 Plus spectrometer (Bruker, Karlsruhe, Germany) equipped with an LC Agilent 1100 Diode-Array Detector (Agilent Technologies, Waldbronn, Germany), using an Ascentis express Supelco RP18, 2.7 μ (50 × 4.6 mm) column kept at 40 °C, eluting at 1 ml min−1 with a 7-min linear gradient from 95:5 phase A (0.05% (v/v) trifluoroacetic acid in water):phase B (0.05% (v/v) trifluoroacetic acid in acetonitrile) to 100% phase B. HR-MS spectra were acquired in fourier transform mode from 400 to 2500 m/z (high-mass range mode, 1 × 106 resolution, automatic gain control (AGC) scan target 5 × 105). Spectra acquired during 30 min were averaged. Samples were infused by syringe at 8 μl min−1 into a LTQ XL Orbitrap (Thermo, West Palm Beach, FL, USA) interfaced with an ESI source. 1H-,13C-, 1- and 2-D NMR spectra (COSY, TOCSY, NOESY, HSQC, HMBC) were measured in CD3CN-D2O at 25 °C using an AMX 600 MHz spectrometer (Bruker). Chemical shifts are reported relative to D2O (δ 4.79) and MeCN. Sequencing of the 16S ribosomal RNA gene was performed following published procedures.23
Actinomycete strains and growth conditions
Actinoplanes strains ID104802 and ID104771 were grown on S1 plates for 10–15 days at 30 °C. S1 (oatmeal, 60 g l−1; agar, 18 g l−1; FeSO4 × 7 H2O, 0.001 g l−1; MnCl2 × 4 H2O, 0.001 g l−1; ZnSO4 × 7 H2O, 0.001 g l−1) was prepared by boiling oatmeal in 1 l distilled water for 20 min, filtering it through cheesecloth, adding the remaining components adjusting volume to 1 l with distilled water and pH to 7.2 with 0.5 M NaOH before sterilization at 121 °C for 20 min. For metabolite production, the microbial strain was scraped from a 90-mm agar plate and inoculated into a 500 ml Erlenmeyer flask containing 100 ml of seed medium (dextrose monohydrate, 20 g l−1; yeast extract, 2 g l−1; soybean meal, 8 g l−1; NaCl, 1 g l−1; calcium carbonate, 4 g l−1; pH 7.3 by 0.5 M NaOH), which was incubated at 30 °C in an orbital shaker set at 200 r.p.m. After 48–72 h, 5 ml of the culture were transferred into 100 ml of fresh medium in a new 500-ml flask. After further 48 h, a 5% inoculum was made into production media M8 (soluble starch, 20 g l−1; glucose, 10 g l−1; yeast extract, 2 g l−1; casein hydrolysate, 4 g l−1; meat extract, 4 g l−1; CaCO3, 3 g l−1; pH 7.2 with 0.5 M NaOH) or KC4 (Pharmamedia) (Traders Protein, Memphis, TN, USA), 30 g l−1; maize dextrin, 40 g l−1; yeast extract, 5 g l−1; glucose monohydrate, 10 g l−1; CaCO3, 2 g l−1; NaCl, 1 g l−1; pH 7.0 with 0.5 M NaOH). For bioreactor experiments a 20-l BioFlo 415 fermenter (New Brunswick Scientific, Enfield, CT, USA) was used, containing 15 l of production medium KC4, with 400 r.p.m. agitation and 0.4-v.v.m. aeration at a temperature of 30 °C. For monitoring metabolite production, methanol extracts were prepared by withdrawing 0.45 ml of culture, thoroughly mixing it with 2 vol methanol:acetic acid 92:8 for 1 h at 50 °C and 1400 r.p.m., followed by centrifugation. The resulting methanol extract was either assayed by HPLC, as described above, or dried for antimicrobial assays, as described below.
Metabolite purification
A 15-l, 72 h culture of strain ID104802 in KC4 medium was filtered through a paper filter and the collected 3 l of mycelium was treated with one volume methanol. The resulting organic phase was concentrated under vacuum to about 1 l and then shaken with 113 ml Diaion HP20 resin (Supelco, Bellefonte, PA, USA) for 3 h at 30 °C. After washing twice with 500 ml 35% MeOH, the resin was eluted with 1.5 l 90% MeOH. After evaporating under vacuum, samples were resuspended in 5 ml of H2O-dimethylformamide 1:1 and purified by reversed-phase medium pressure liquid chromatography on a Combiflash system (Teledyne ISCO, Lincoln, NE, USA), using an RP18 86 g column, a 20-min linear gradient of 0.05 M ammonium formate-MeOH (from 35 to 95% eluent B) at 60 ml min−1 with a 214-nm detection wavelength. Fractions eluted with 85% eluent B, which exhibited activity against S. aureus ATCC 6538P, were combined and, after removing methanol under vacuum, were lyophilized, affording 907 mg 1. The broth filtrate obtained from the fermentation run was also absorbed onto the HP20 resin (using 10 g l−1) and processed as above, yielding 568 mg 1.
Chemistry
Edman degradation: One milligram of 1 was dissolved in 500 μl of 1 M Na2CO3 (pH 8) and phenylisothiocyanate (1 μl) was added. The reaction was stirred at 60 °C for 1 h before MS analysis. Amino acid composition analysis: 1 (3 mg) was hydrolyzed in 1 ml 6 M HCl at 160 °C for 5 min under microwave irradiation. The hydrolyzed sample was evaporated to dryness, resuspended in 1 ml H2O-acetonitrile 1:1 and treated with 4-(4-isothiocyanate-phenyl)-azo-N,N-dimethyl aniline (5 mg) and Triethylamine (6 μl). The reaction mixture was stirred for 2 h at 60 °C and then extracted twice with petroleum ether: dichloromethane 8:2 (3 ml). The organic phase was evaporated to dryness, redissolved in 1 ml water-acetonitrile 1:1 and analyzed by HPLC-MS. Ethanethiol derivatization (alkaline pH):9 One milligram 1 was dissolved in 200 μl dimethylformamide and then 200 μl of the derivatization mixture were added (780 μl ethanol, 560 μl water, 180 μl of 5 M NaOH and 168 μl ethanethiol). The mixture was kept for 1 h at 60 °C and then directly analyzed by LC-MS. Ethanethiol derivatization (neutral pH):8 1 mg 1 was dissolved in acetonitrile - 0.1 M ammonium acetate 1:1 (800 μl) and then 3 μl of ethanethiol were added. The mixture was kept for 1 h at 60 °C and then directly analyzed by LC-MS. Amidation with benzylamine: to a stirred solution of 2 mg 1 in 1 ml dimethylformamide, 2 μl of benzylamine (20 Eq.) and 9.4 mg PyBOP (20 Eq.) were added; the reaction was kept under stirring for 20 min at room temperature before LC-MS analysis. Amidation with ethylenediamine: to a stirred solution of 30 mg 1 in 2 ml dimethylformamide, 5.5 μl of ethylendiamine (6 Eq.) and 16.5 mg of PyBOP (2 Eq.) were added and the reaction was kept under stirring for 20 min at room temperature. The reaction product was purified on a 4.3 g reverse-phase C18 RediSep Column (Teledyne ISCO) by using the Combiflash System. The resin was eluted at 18 ml min−1 with a linear gradient from 5 to 90% of phase B in 18 min. Phase A was 0.05 M ammonium formate whereas phase B was acetonitrile. After LC-MS analysis fractions containing the monoamide were pooled, concentrated under vacuum and lyophilized, yielding 12 mg of 3.
Physico-chemical properties
NAI-802 (1): white powder, UV (in CH3CN:H2O=50:50 with 0.05 % of trifluoroacetic acid) 225, 280 nm. 1H- and 13C-NMR (600 MHz, CD3CN-D2O) (Table 1 and Supplementary information). (+)-HR-ESI MS (m/z) 1058.9707 [M+2H]2+, 706.3151 [M+3H]3+ calculated for C90H143N25O26S4 1058.9754 [M+2H]2+. MS/MS fragmentation (Table 2).
Ala(0) NAI-802 (2): white powder, UV (in CH3CN:H2O=50:50 with 0.05 % of trifluoroacetic acid) 225, 280 nm. (+)HR-ESI MS (m/z) 1094.4889 [M+2H]2+ 729.9937 [M+3H]3+ calculated for C93H148N26O27S4 1094.4940 [M+2H]2+. MS/MS fragmentation (Table 2).
NAI-802 monoamide with ethylenediamine (3): white powder, UV (in CH3CN:H2O=50:50 with 0.1 % of trifluoroacetic acid) 225, 280 nm. (+)HR-ESI MS (m/z) 1080.0120 [M+2H]2+, 720.3453 [M+3H]3+ calculated for C92H149H27O25S4 1080.0045 [M+2H]2+. MS/MS fragmentation (Table 2).
Antimicrobial assays
After drying, methanol extracts or purified fractions were resuspended in 100 μl 10% dimethyl sulfoxide and 20 μl were deposited on 4-mm thick Müller Hinton Agar (Difco Laboratories, Detroit, MI, USA) plates, inoculated with 105 c.f.u. ml−1 S. aureus ATCC 6538P. Plates were incubated overnight at 37 °C before scoring inhibition halos.
MIC assays were performed by the broth microdilution methodology in sterile 96-well microtiter plates according to Clinical and Laboratory Standards Institute (CLSI) guidelines;24 media used were Müller Hinton Broth containing 20 mg l−1 CaCl2 and 10 mg l−1 MgCl2 for all strains, except for Streptococcus spp. and C. albicans, which were grown in Todd Hewitt Broth (Difco Laboratories) and in RPMI 1640 (Sigma Aldrich, St Louis, MO, USA) supplemented with 0.165 M MOPS (pH 7), respectively. When indicated, bovine serum was added to the medium at 30% (w/v). Bacteria were inoculated at 5 × 105 c.f.u. ml−1, while C. albicans at 5 × 104 c.f.u. ml−1. After 24-h incubation (48 h for C. albicans) at 37 °C, MIC was defined as the lowest drug concentration causing complete suppression of visible growth. Compounds were dissolved in dimethyl sulfoxide except for vancomycin, which was dissolved in water. Appropriate dilutions were made with the required culture medium immediately before testing.
MICs for anaerobic bacteria were determined by the broth dilution method in Brucella broth supplemented with 5 μg ml−1 hemin, 1 μg ml−1 vitamin K1, 5% lysed horse blood and 1:25 (v/v) Oxyrase (Mansfield, OH, USA), as described.25 Inocula were prepared by suspending few colonies from a 48-h agar plate in Brucella broth to an OD625 of 0.8, then diluting 1:10 with Brucella broth to achieve a final titer of about 105 CFU ml−1. Cultures were incubated for 48 h at 37 °C under 80% N2, 10% CO2 and 10% H2, using a GasPak EZ anaerobe container system (Becton Dickinson, Buccinasco, Milano, Italy).
References
Castiglione, F. et al. Determining the structure and mode of action of Microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem. Biol. 15, 22–31 (2008).
Jabes, D., Brunati, C., Guglierame, P. & Donadio, S. In vitro antibacterial profile of the new lantibiotic NAI-107. Abstracts of 49th Intersci Conf on Antimicrob Agents Chemother, No. F1-1502 ( San Francisco, (2009).
Jabes, D. et al. Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob. Agents Chemother. 55, 1671–1676 (2011).
Jabes, D. & Donadio, S. Strategies for the isolation and characterization of antibacterial lantibiotics. Meth. Mol. Biol. 618, 31–45 (2010).
Lazzarini, A., Cavaletti, L., Toppo, G. & Marinelli, F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie Van Leeuwenhoek 79, 399–405 (2000).
Zimmermann, N., Metzger, J. W. & Jung, G. The tetracyclic lantibiotic actagardine 1H-NMR and 13C-NMR assignments and revised primary structure. Eur. J. Biochem. 228, 786–797 (1995).
Wüthrich, K. NMR of Proteins and Nucleic Acids, Wiley: New York, NY, (1986).
Smith, L. et al. Covalent structure of mutacin 1140 and a novel method for the rapid identification of lantibiotics. Eur. J. Biochem. 267, 6810–6816 (2000).
Meyer, H. E. et al. Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal. Biochem. 223, 185–190 (1994).
Vertesy, L. et al. Ala(0)-actagardine, a new lantibiotic from cultures of Actinoplanes liguriae ATCC 31048. J. Antibiot. 52, 730–741 (1999).
Cotter, P. D., Hill, C. & Ross, R. P. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Peptide Sci. 6, 61–75 (2005).
Willey, J. M. & van der Donk, W. A. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501 (2007).
Schneider, T. & Sahl, H. G. An oldie but a goodie – cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 300, 161–169 (2010).
Velásquez, J. E. & van der Donk, W. A. Genome mining for ribosomally synthesized natural products. Curr. Opin. Chem. Biol. 15, 11–21 (2011).
Somma, S., Merati, W. & Parenti, F. Gardimycin, a new antibiotic inhibiting peptidoglycan synthesis. Antimicrob. Agent Chemother. 11, 396–401 (1977).
Boakes, S., Appleyard, A. N., Cortés, J. & Dawson, M. J. Organization of the biosynthetic genes encoding deoxyactagardine B (DAB), a new lantibiotic produced by Actinoplanes liguriae NCIMB41362. J. Antibiot. 63, 351–358 (2010).
Holtsmark, I., Mantzilas, D., Eijsink, V. G. H. & Brurberg, M. B. Purification, characterization, and gene sequence of Michiganin A, an Actagardine-like lantibiotic produced by the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Appl. Environ. Microbiol. 72, 5814–5821 (2006).
Boakes, S., Cortés, J., Appleyard, A. N., Rudd, B. A. & Dawson, M. J. Organization of the genes encoding the biosynthesis of actagardine and engineering of a variant generation system. Mol. Microbiol. 72, 1126–1136 (2009).
Cooper, L. E., McClerren, L., Chary, A. & van der Donk, W. A. Structure-activity relationship studies of the two-component lantibiotic Haloduracin. Chem. Biol. 15, 1035–1045 (2008).
Ross, A. C. & Vederas, J. C. Fundamental functionality: recent developments in understanding the structure-activity relationships of lantibiotic peptides. J. Antibiot. 64, 27–34 (2011).
Malabarba, A., Pallanza, R., Berti, M. & Cavalleri, B. Synthesis and biological activity of some amide derivatives of the lantibiotic actagardine. J. Antibiot. 9, 1089–1097 (1990).
Donadio, S., Maffioli, S., Monciardini, P., Sosio, M. & Jabes, D. Sources of novel antibiotics--aside the common roads. Appl. Microbiol. Biotechnol. 88, 1261–1267 (2010).
Mazza, P., Monciardini, P., Cavaletti, L., Sosio, M. & Donadio, S. Diversity of Actinoplanes and related genera isolated from an italian soil. Microb. Ecol. 45, 362–372 (2003).
National Committee for Clinical Laboratory Standards Performance standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement – CLSI document M100-S20, NCCLS, Wayne, Pennsylvania, USA (2010).
National Committee for Clinical Laboratory Standards.. Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria; Approved Standard – Sixth Edition, 2004 – NCCLS document M11-A6, NCCLS, Wayne, Pennsylvania 19087-1898 USA (2004).
Acknowledgements
This work was partially supported by grants from Italian MIUR and Regione Lombardia. MS was directly supported from a fellowship from Regione Lombardia. We are grateful to Vincenzo Rizzo and Francesca Vasile (Center of biomolecular Interdisciplinary Studies and Industrial applications) for their expert advices on analytical techniques.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Simone, M., Monciardini, P., Gaspari, E. et al. Isolation and characterization of NAI-802, a new lantibiotic produced by two different Actinoplanes strains. J Antibiot 66, 73–78 (2013). https://doi.org/10.1038/ja.2012.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2012.92
Keywords
This article is cited by
-
Actinoplanes aureus sp. nov., a novel protease-producing actinobacterium isolated from soil
Antonie van Leeuwenhoek (2021)
-
Advancing cell wall inhibitors towards clinical applications
Journal of Industrial Microbiology and Biotechnology (2016)
-
Growing the seeds sown by Piero Sensi
The Journal of Antibiotics (2014)