Abstract
Taxonomic classification of the thousands–millions of 16S rRNA gene sequences generated in microbiome studies is often achieved using a naïve Bayesian classifier (for example, the Ribosomal Database Project II (RDP) classifier), due to favorable trade-offs among automation, speed and accuracy. The resulting classification depends on the reference sequences and taxonomic hierarchy used to train the model; although the influence of primer sets and classification algorithms have been explored in detail, the influence of training set has not been characterized. We compared classification results obtained using three different publicly available databases as training sets, applied to five different bacterial 16S rRNA gene pyrosequencing data sets generated (from human body, mouse gut, python gut, soil and anaerobic digester samples). We observed numerous advantages to using the largest, most diverse training set available, that we constructed from the Greengenes (GG) bacterial/archaeal 16S rRNA gene sequence database and the latest GG taxonomy. Phylogenetic clusters of previously unclassified experimental sequences were identified with notable improvements (for example, 50% reduction in reads unclassified at the phylum level in mouse gut, soil and anaerobic digester samples), especially for phylotypes belonging to specific phyla (Tenericutes, Chloroflexi, Synergistetes and Candidate phyla TM6, TM7). Trimming the reference sequences to the primer region resulted in systematic improvements in classification depth, and greatest gains at higher confidence thresholds. Phylotypes unclassified at the genus level represented a greater proportion of the total community variation than classified operational taxonomic units in mouse gut and anaerobic digester samples, underscoring the need for greater diversity in existing reference databases.
Similar content being viewed by others
Introduction
In medical and environmental microbiome studies that employ high-throughput (HTP) sequencing of 16S rRNA genes, the Roche 454 and Illumina platforms have largely supplanted traditional Sanger sequencing (Margulies et al., 2005). Barcodes unique for each sample, and added to sequences in the PCR step, allow the multiplexing of hundreds or more samples per instrument run, enabling robust study designs to be used economically in place of anecdotal descriptions of microbiota (Binladen et al., 2007; Hoffmann et al., 2007; Huber et al., 2007; Hamady et al., 2008; McKenna et al., 2008). Taxonomic classification is a critical and informative component of the many software pipelines used by researchers to probe community structure using bacterial 16S rRNA gene sequence data. Furthermore, taxonomic classification informs and complements results from studies using techniques from classical ecology, especially relative abundance measurements, diversity measurements and ordination. For instance, relative abundance measurements are obtained by assigning 16S rRNA reads to similar clusters (that is, operational taxonomic units, or OTUs), either by homology methods such as basic local alignment search tool (Altschul et al., 1990) and UCLUST (Edgar, 2010), or by shared occurrence of short oligonucleotides (Wang et al., 2007), and interpretation of the relative abundances is then informed by assignments of taxonomic classification to the OTUs.
There are many taxonomic classification algorithms available (Liu et al., 2008), and several reference databases that can be used with any given algorithm. Due to the large number of sequences obtained from HTP 16S rRNA gene sequencing-based studies, a commonly employed practical approach for taxonomic classification is the naïve Bayesian method developed for the RDP (Cole et al., 2009) by Wang et al (Wang et al., 2007). This algorithm has proven its utility and has sustained considerable popularity since its introduction. Liu et al. (2008) performed an extensive survey of different classification methods, and concluded that naïve Bayesian RDP Classifier and the Simrank (DeSantis et al., 2011) and DNADIST (Felsenstein, 1989) search approach on the Greengenes (GG) website were the two most useful and informative methods for classifying HTP 16S rRNA gene sequences. A naïve Bayesian classification method builds a statistical model from a list of all words of a given length (RDP uses 8-mers as default) present in a training set, and classifies a query sequence based on the probability that randomly selected words appear at different nodes in the taxonomic mapping of the training set. The training set consists of two sets of data: a database of reference sequences, and a taxonomic hierarchy mapped to each of the reference sequences. Both the specific sequences used as references, and the specific taxonomy applied to them, may affect classification results.
As research groups explore different custom classification training sets for in-house 16S rRNA gene sequence processing, one factor to consider is the gene region sequenced. Due to the limits on sequence length in next generation sequencing platforms, a hypervariable region of the 16S rRNA gene (Neefs et al., 1993) and its corresponding primer pairs must be selected. Huse et al. (2008) used their alignment-based GAST algorithm to demonstrate that pyrosequencing reads from the V1 to V2 and V6 regions are useful proxies for full-length sequences when performing taxonomic classification. For the naïve Bayesian algorithm, however, the relative positions of 8-mer words are lost when building a classification model. This presents the possibility that a word from the primer-targeted region of the query sequence may match, by random chance, a word from a different region of a full-length reference sequence, especially for words of low complexity. It is unclear to what extent this sequence noise in the reference database may impact classification results.
In this study, our goal was to compare naïve Bayesian taxonomic classification results using training sets built from three different reference databases of varying diversity and overall taxonomic structure. We applied the training sets to classify sequences generated from five different studies, including samples from different human body locations, mouse gut, python gut, soil samples and anaerobic digester sludge. We tested the different training sets in their original sizes, and also generated versions where sequence count was standardized. To test whether differences in training set performance were due to differing sequence content or differing taxonomies, we tested different training sets that shared a single taxonomy and different taxonomies for the same reference sequences. We also tested the effect of sequence noise in the reference database by trimming sequences to the primer region before training the classification model. Finally, we asked how unclassified OTUs generated with the different training sets were distributed phylogenetically, and how well classified and unclassified OTUs represented the β-diversity within studies. Our results suggest that researchers using in-house pipelines for sequence processing would benefit from including as much diversity as possible in their reference database for taxonomic classification. Additionally, trimming the reference sequences to the primer region of the query sequences affords significant benefits to naïve Bayesian classification tasks, especially when a high-confidence threshold is desired. Because classified OTUs can, in some studies, represent less of the β-diversity than unclassified OTUs, the latter should not be removed before from β-diversity measures.
Materials and methods
Sequence processing
To compare taxonomic classification results using microbiome data sets from a wide range of source environments, we chose five published bacterial 16S rRNA gene pyrosequencing data sets: (1) a study of the human microbiome by Costello et al. (2009), which included samples from gut, skin, oral cavity, external auditory canal, nostril and hair (ERA000159); (2) a study of the mouse gut microbiome by Ravussin et al. (2011; SRA022795); (3) a time series of a python gut microbiome throughout a feeding cycle by Costello et al. (2010; SRA012490); (4) a survey of a diverse range of soil samples by Lauber et al. (2009); (5) a time series of nine upflow anaerobic digesters by Werner et al. (2011; SRA029112). All data sets that we chose for this study were sequenced via barcoded 454 pyrosequencing (FLX chemistry) of 16S rRNA genes using the bacterial primers for the V1–V2 region (8F-338R), as described by Hamady et al. (2008).
For each of the sequencing data sets, raw SFF files and sample mapping files (that is, files that relate each unique barcoded primer sequence to its associated sample) were acquired from the authors and processed using the default settings in the Quantitative Insights Into Microbial Ecology pipeline (Caporaso et al., 2010b). Sequences were filtered to exclude low-quality reads and primer/barcode regions were trimmed. Flowgrams were denoised using Denoiser 0.84 (Reeder and Knight, 2010), and clustered into OTUs at 97% pairwise identity (ID) using the UCLUST (Edgar, 2010) seed-based algorithm. A representative sequence from each OTU was aligned to the GG-imputed core reference alignment (DeSantis et al., 2006) using PyNAST (Caporaso et al., 2010a), and the concatenated alignment of OTUs was filtered to remove gaps and hypervariable regions using the GG Lane mask (DeSantis et al., 2006). A phylogenetic tree was constructed from the filtered alignment using the approximately maximum likelihood algorithm implemented in FastTree (Price et al., 2010). An unweighted UniFrac distance matrix (Lozupone and Knight, 2005) was constructed from the phylogenetic tree, and phylogenetic β-diversity was visualized by applying principal coordinates analysis to the UniFrac distance matrix.
Taxonomic classification
All taxonomic classifications were assigned using the naïve Bayesian algorithm (Wang et al., 2007) developed for the RDP classifier (Cole et al., 2009), as implemented in Mothur 1.15 (Schloss et al., 2009). For all analyses, we ran the classifier using a confidence threshold value of 80%, unless otherwise specified. To build a naïve Bayesian model for taxonomic classification requires a training set, which consists of a database of reference sequences and a taxonomy file assigning taxonomic hierarchy to each sequence. The training sets we tested for naïve Bayesian taxonomic classification were obtained from established-sequence-processing pipelines (Table 1). RDP Training Set 6 (RDP TS6) was the default in the latest RDP classifier 2.2 (Cole et al., 2009); a subset of the SILVA database (SILVA subset) was the default training set distributed for the Mothur software package (Schloss et al., 2009). Additionally, to test the advantages of greater phylogenetic diversity in the training set, we built a new training set from the full GG database and the latest GG taxonomy (GG99; described below). The GG99 training set was significantly larger (127 741 sequences) than RDP TS6 (8422 sequences) or the SILVA subset (14 956 sequences). To determine the effect of unclassified OTUs on the total observed phylogenetic variation, the OTU table (relating sequence counts for each OTUs to the samples they were obtained from) from each original data set was split into two separate OTU tables based on GG classification results: (1) OTUs successfully classified at the genus level, and (2) OTUs that were unclassified at the genus level, resulting in three total OTU tables per data set (the whole OTU table, plus classified and unclassified OTU tables). Unweighted UniFrac analysis and principal coordinates analysis was performed, as described above, using each of the classified and unclassified OTU tables to subsample the phylogenetic tree.
GG classification subset
The subset of unique sequences from GG (GG99) was selected to train the classifier from the GG database (full, unaligned, 21 May 2010) and the latest GG taxonomy (17 December 2010). Reference sequences that were assigned a classification in the GG taxonomy were extracted from the GG database, and a representative subset of those sequences was selected by clustering at 99% ID using UCLUST (Edgar, 2010). The GG taxonomy was based on a tree generated with FastTree (Price et al., 2010) and inferred from an Infernal alignment (Nawrocki et al., 2009) of 408 135 chimera-filtered (Chimera Slayer; Haas et al., 2011) sequences in the GG database. Taxonomic informative classifications available for a subset of deposited 16S rRNA sequences (mostly isolates) were placed on the inferred tree using a sensitivity/specificity optimization and propagated to all sequences to produce the final GG taxonomy. The GG taxonomic hierarchy specifies taxa only at levels for which there was evolutionary branching of reference sequences to support the designation of separate lineages.
To independently determine the effect of training set size on classification results, we reduced the size of the larger training sets to match the size of the smallest set (RDP TS6) by clustering sequences at lower levels of ID. Representative sequences were picked using the default Quantitative Insights Into Microbial Ecology pipeline. We used a subset of GG clustered at 91.3% ID (GG91.3; 8275 sequences; Table 1) and a subset of SILVA clustered at 98.7% ID (SILVA98.7; 8572 sequences; Table 1). We also mapped the GG taxonomy to the full set of reference sequences in the RDP TS6, for a direct comparison in which the taxonomic hierarchy changed but the reference sequences remained the same.
To test the effect of trimming the reference database to match the primer region, we created two additional training sets from the GG database. We trimmed GG in-silico in the backward direction from the 338R position, and then clustered at 99% ID, as the first of these additional training sets. Due to differential clustering of trimmed training set sequences, for direct comparison between trimmed and untrimmed training sets, a second training set consisting of full-length versions of the successfully trimmed and clustered GG sequence representatives was used.
Results and discussion
Training set choice influences taxonomic abundances
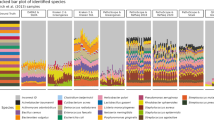
Compared with RDP TS6, the GG99 training set contained the same major phyla represented and a similar overall range of pairwise distances. However, GG99 included a much greater extent of diversity within each phylum than the other training sets tested, as observed from a pattern of denser clouds of OTUs from GG versus RDP TS6 in principal coordinates analysis plots of pairwise sequence distances (Supplementary Figure S1). We compared the distribution of sequences across classes for the different databases: GG99 included a greater number of taxonomic classes than RDP TS6 and the SILVA subset. Additionally, GG99 contained multiple unique representative sequences for each class, and many classes with >10 sequences per class, whereas RDP TS6 and the SILVA subset contained several classes with only one sequence representative, and many more with 10 or fewer representative sequences (Supplementary Figure S2). These observations indicate that the greater sequence count of GG99 was distributed throughout the taxonomic divisions. When left in their original size, the three training sets yielded varying relative abundances of the major phyla identified, depending on which of the query data sets were analyzed (Figure 1). For instance, when analyzing the human gut sequences (Figure 1a), the RDP TS6 and the SILVA subset training sets were unable to classify 6% of the sequences, whereas GG99 was unable to classify only 1% of the sequences.
Relative abundance of the 10 major phyla identified by naïve Bayesian classification using five different training sets: three of approximately the same size: GG91.3, SILVA98.7, and RDP TS6, and two larger training sets: GG99 and the SILVA subset for Mothur. Relative abundances were averaged for samples of five different studies (note that human gut is shown apart from non-gut samples from the sample study): (a) human gut, (b) non-gut human body locations, (c) mouse gut, (d) anaerobic digester, (e) python gut and (f) soils.
The use of different training sets resulted in different relative abundances of specific phyla, notably the Synergistetes, Spirochetes, Tenericutes and Chloroflexi, which all had greater relative abundances based on the GG99 training set for several of the data sets (Figure 1). Furthermore, sequences that were assigned to these phyla using GG99 accounted for a significant portion of the sequences that were unclassified by RDP TS6 or the SILVA subset. The gut samples (human, mouse and python) in particular contained sequences that were classified as Tenericutes only with the GG99 training set. In the human gut set, GG99 classified 38% of the sequences as Firmicutes and 1% as Tenericutes, whereas RDP TS6 and the SILVA subset failed to classify the Tenericutes and classified 35% and 36% of all sequences as Firmicutes, respectively. Although the RDP TS6 training set includes a Tenericutes phylum, the GG99 training set had a distinctively higher diversity of Tenericutes (1430 entries in GG99, compared with 170 in RDP TS6), which may account for why GG99 classified more sequences to this phylum. Similarly to the human gut samples, the mouse cecal samples contained sequences that GG99 classified as Tenericutes but that RDP TS6 and the SILVA subset did not (Figure 1c).
Other discrepancies in classification outputs were specific to the different studies. For instance, in the mouse-gut data set, one highly abundant OTU was classified using GG99 as Allobaculum, but was unclassified using RDP TS6 and the SILVA subset. As a result, 50% of the mouse gut sequences were unclassified by RDP TS6 and the SILVA subset compared with 21% unclassified by GG99. For the python-gut data (Figure 1e), RDP TS6 and the SILVA subset did not classify any Tenericutes, and GG99 found 1% of the sequences to belong to that phylum. RDP TS6 and GG99 classified 4% of the sequences as Synergistetes, in contrast to the SILVA subset, which did not classify any sequences to this phylum. Discrepancies between training sets for the soil data set involved other phyla as well: more than 25% of the soil sequences were classified as Acidobacteria by RDP TS6 and GG99 compared with 20% by the SILVA subset (Figure 1f). RDP TS6 failed to classify the soil Chloroflexi that were found with the GG99 and the SILVA subset training sets. The anaerobic digester samples (Figure 1d) contained significant Spirochetes populations that were identified using the SILVA subset and GG training sets, but not with RDP TS6.
For generally more numerically dominant phyla (Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria and Acidobacteria), relative abundance results agreed well among the three full-size training sets used. In the human gut samples, for example, all three training sets classified 54% of the sequences as Bacteroidetes and 4% as Proteobacteria. This lends support to the use of alternative SILVA subset and GG99 training sets for classification, because similar results were obtained in comparison with the well-established RDP TS6.
Performance relates to training set size
To control for the effect of size (sequence count) of the training sets on their performance, the two larger training sets were reduced to the size of RDP TS6 (see Materials and methods): GG99 yielded the smaller GG91.3 and the SILVA subset yielded SILVA98.7 (Table 1). The phylogenetic depth of the classification results (that is, phylum, class, order and so on) increased systematically as a function of training set size (Supplementary Figure S3). Furthermore, for all five data sets tested, the number of OTUs classified was systematically reduced as a function of training set size. For example, on average, roughly 25% of genus-level IDs were lost to the unclassified category when GG was reduced from 127 741 sequences to 8275 (Supplementary Figure S4). Overall, performances were similar between the similarly sized training sets, with some important differences. For instance, for the mouse-gut data, the GG performance dropped with the reduction in size (Figure 1c). This is probably due to the loss of training set sequences that were key to the classification of a subset of the mouse gut sequences (mostly Proteobacteria). However, Tenericutes were still classified by GG91.3, despite its smaller size, while they were not classified by the other training sets. Supplementary Figure S4 also suggests that clustering a training set at 97% ID, instead of 99% ID, results in an insignificant loss in classification precision. We therefore suggest that it would be practical for users to classify sequences using the 97% ID GG OTU database available for download (http://greengenes.lbl.gov/Download/Sequence_Data/Fasta_data_files/Reference_OTUs_for_Pipelines).
Little effect of taxonomic hierarchy differences between training sets
To check for the effect of differing taxonomic hierarchies on classification outcomes, we reduced GG99 to include only the sequences contained in the RDP TS6 training set. Thus, for this comparison, the training sets differed only by their taxonomic hierarchies but had identical sequence sets. Wang et al. (2007) pointed out that the RDP taxonomy, based on Bergey's taxonomy, was not built by parsing of genetic phylogeny, and it is known to contain some errors. However, our results comparing different taxonomies for the same training set reference sequences suggest that, overall, both the RDP taxonomy and the GG taxonomy yield similar classification precision, on average. In other words, differences in taxonomic hierarchies were less important than differences in sequence diversity for explaining discrepancies between outcomes. Compared with using the RDP taxonomy, use of the GG taxonomy did not result in statistically significant differences in the number of OTUs classified at most levels of taxonomy, except for marginal improvements at the genus level (Supplementary Figure S5). Genus-level differences were attributed to how reference sequences were annotated with taxonomic information; because the full GG database contains a greater number of sequence representatives in different taxonomic groups, the GG taxonomic annotations are deeper for sets of sequences that, in the RDP TS6 training set, are the sole representatives of higher-order taxonomic groups (for example, classes, orders), and which are therefore not annotated as deeply in RDP TS6, to avoid giving misleadingly specific taxonomic information when a query sequence falls within those groups. Differences between taxonomic hierarchies may impact abundance-based results in specific (and probably rare) cases where an OTU is both abundant in the sample and happens to be classified differently in training sets.
OTUs with low-classification depth clustered by evolutionary history
To gain further insight into the effects of training sets, we additionally assessed the results of different classification training sets on individual OTUs, irrespective of their relative abundances in the data sets. Figure 2 summarizes classification depth for each OTU from both the human body (Figure 2a) and the soil (Figure 2b) sequences. Similar plots of the mouse gut, python gut and anaerobic digester sequences are available in Supplementary Figure S6. Heat maps representing classification depth for all OTUs were mapped onto phylogenetic trees.
Summary of OTU classification depth using each of the three training sets for two of the four studies: (a) human body OTUs, and (b) soil OTUs (other three data sets shown in Supplementary Figure S6). OTUs are organized according to evolutionary history, as determined by the FastTree approximately-maximum-likelihood tree constructed in the default QIIME pipeline. Inset charts summarize the total number of OTUs classified at each taxonomic level (GG99=dark blue, GG91.3=light blue, SILVA=green, RDP TS6=orange).
Classification failures were phylogenetically clustered, rather than distributed randomly (Figure 2, Supplementary Figure S6). This suggests that unclassifiable OTUs could be attributed to the presence of distinct phylogenetic lineages in the query samples that were not represented well in the training sets. The effect of these underrepresented lineages was especially evident in the soil samples, which contained large groups of poorly classified sequences belonging to the Actinobacteria, Chloroflexi, Deltaproteobacteria, Armatimonadetes (previously OP10), TM6 and TM7, among others. Because the trees in Figure 2 are based on a small region of the 16S rRNA gene, higher order evolutionary branching was not consistently resolved compared with the GG phylogeny and associated taxonomy, which was based on full-length sequences. However, the clusters of highly similar sequences were taxonomically consistent, and informative of the evolutionary relationships between similar OTUs.
A bias of known reference sequences toward representing the human microbiome is visually evident when comparing the two heat maps in Figure 2, especially when comparing individual phyla. For example, Actinobacteria OTUs in the human body samples had extensive genus- and species-level coverage using all of the training sets, compared with soil samples in which Actinobacteria OTUs (and close relatives of unknown classification) had groups of poor classification depth via all of the training sets. In human body samples, the most evident groups with poor representation in the training sets were TM7 and Chloroflexi. The mouse gut classification results (Figure 1c) underscore the need for greater representation of Tenericutes in training sets used to study mammalian gut microbiomes.
Trimming training set sequences improves classification results
We next tested the effect on classification results of restricting the length of the sequences in the training sets to the 16S rRNA gene amplicon region generated in the studies. All the data sets we considered in this study were sequenced using the universal bacterial primers spanning the V1–V2 region, so no experimental sequence information was available beyond the E. coli 16S rRNA position 338. The reference sequences in the classifier training sets, however, were full length. We tested the hypothesis that including the reference sequence information beyond the 338R position would contribute noise to the classification results.
Trimming the training set always improved classification depth, for all sample origins, at all percentage confidence threshold values, and at all taxonomic levels (Figures 3a and b). For example, for classification with 80% confidence, the number of OTUs classified at the genus level improved by 6–14%, depending on the sample type (Figure 3c). We tested whether the gains realized from trimming the reference sequences would be greater when a more stringent confidence threshold was applied. Aside from altering the content and size of the training set, the confidence threshold is another important parameter of the model. In naïve Bayesian classification, the confidence threshold is the minimum consensus support required to assign taxonomy at any given level (for example, if the model is applied 100 times with a confidence threshold of 80%, then a consensus taxonomy must appear 80/100 times for it to be assigned by the model). As expected, classification with higher threshold (60%, 80% or 95%) did indeed yield greater improvements upon trimming in the percentage of classified OTUs (Figure 3c), as well as a greater total increase in the number of classified OTUs (Figure 3b). The only exception was at the genus and species levels, where trimming yielded similar total gains at all confidence thresholds, with some variation depending on the sample origin (Figure 3b). These results demonstrate that the decision to trim the reference database improved all classification queries, and had a greater impact for analyses in which a higher confidence threshold was desired.
The effect of trimming the GG99 training set on classification depth, for each of the five data sets: (a) the total number of OTUs classified at each taxonomic level (Tr=trimmed; full=full length; color key indicates different percentage confidence thresholds applied to the naïve Bayesian model: 60%, 80% and 95%), (b) the total number of classified OTUs gained as a consequence of trimming the training set and (c) the percentage gain of total OTUs classified as a consequence of trimming the training set. Note that all values in (b) and (c) are positive, indicating that trimming always afforded a net gain in classification precision.
Unclassifiable OTUs captured a significant portion of the total phylogenetic variation
We assessed the impact of classified and unclassified OTUs on downstream analysis. Due to the phylogenetic clustering of unclassified OTUs (Figure 2), we expected that removal of unclassified OTUs from an experimental data set would impact measures of β-diversity. To test this, we compared the ability of OTUs that were either classifiable or unclassifiable at the genus level with explain the overall microbial community variation. There are a number of ways to quantify community variation and β-diversity, in addition to taxonomic summaries. These include statistical comparisons of OTU relative abundance profiles, as well as the UniFrac comparison of phylogenetic structure. UniFrac, which was our chosen β-diversity metric for this comparison, quantifies the fraction of total evolutionary history represented in a pair of samples that is unique to one of the samples (Lozupone and Knight, 2005). A UniFrac distance thereby captures information about the phylogenetic dissimilarity of different samples. We performed UniFrac analysis on the whole sequence set for each of the five studies, as well as separate analyses on OTUs that were either classified or unclassified at the genus level by the GG99 training set.
The results of this analysis differed for each data set used and yielded some surprising results. For all sample types, OTUs that were not classifiable at the genus level were important components in the β-diversity, and of the overall phylogenetic structure. For example, in soil samples, OTUs classified to the genus level explained the overall community variation well (Figure 4a; R2=0.984±0.003). However, the GG99 training set failed to classify soil OTUs that were useful in describing phylogenetic variation: UniFrac principal coordinate 1 for unclassified OTUs also correlated well with principal coordinate 1 of the full data set (Figure 4b; R2=0.964±0.008). Indeed, when we compared the results from all the five data sets (UniFrac principal coordinates analysis data shown in Supplementary Figure S7), unclassified OTUs represented a significant contribution to the overall β-diversity, with correlation coefficients (R2) ranging from 0.85 to 0.96 for subsets of unclassified OTUs (Figure 4c). And furthermore, in the mouse gut and anaerobic digester samples, the unclassifiable OTUs were better representatives of overall β-diversity compared with the successfully classified OTUs. These results suggest that, depending on the nature of the sample and its origin, strategies for filtering sequencing results based on the ability to classify the query sequences may result in loss of informative data. Additionally, the use of relative abundances of classified OTUs alone, omitting unclassified OTUs, to characterize community structure may miss out on important components of the microbial community, especially in anaerobic systems.
Ability of either classified or unclassified OTUs alone to recapture the variance of the whole data set. OTUs were classified using the GG99 training set. Each of the plots represent the phylogenetic variation among all soil samples, calculated using all OTUs, along the x axis (UniFrac principal coordinate 1; PC1), correlated to either OTUs that were classified to the genus level (a) or to unclassified OTUs at the genus level (b), on the y axis. If the variance in the OTU subsets (classified or unclassified) explains as much variance as the whole set, then a straight diagonal line is expected. The summary of R2 values for a similar analysis of each of the five sample types is shown for comparison (c). Error bars, and errors on R2 values, represent the s.d. of 10 rarefactions, 200 sequences each. Soil sample data are shown in (a) and (b); other four UniFrac data sets available in Supplementary Figure S7.
Prospectus: choosing a classification training set
We have assessed the advantages of greater training set diversity (higher number of unique sequences) and specificity (trimmed versus untrimmed) for naïve Bayesian taxonomic classification of HTP 16S rRNA gene sequences. The size of the training set had only a slight impact on the computational resources required. For example, we used one 3.0 GHz processor core (without parallel processing) on a Mac OS × system with 16 GB memory, and the model built on the GG99 training set classified 21±5 sequences per computer processing unit (CPU)-second (seq per CPU s), compared with 62±12 seq/CPU s using either the RDP TS6 or SILVA subset training sets. The larger GG99-based classification model occupied 1.3 GB RAM, compared with 0.5 GB RAM for either RDP TS6 or the SILVA subset. The trimmed GG99 training set, although built from shorter reference sequences, resulted in a classification model with CPU and memory needs that were not improved (18±5 seq per CPU s and 1.4 GB RAM) compared with the full-length training set. Based on these computational requirements, the GG99-based classification model can easily be implemented for in-house applications, though it may be impractical to offer as an online service. For future applications, the benefits of trimming must be weighed against other factors, including the success rate for in-silico trimming at the region in question, and the practicality of amassing and managing numerous different classification models for studies in which different regions were sequenced.
The sample origin and the desired consistency with previous measurements are also factors to consider when choosing a training set. Our results have shown that, for generating broad summaries of human body microbiomes, each of the training sets performed well. For example, if taxonomic comparisons with previous human microbiome studies are desired, one might choose RDP TS6 for simplicity and agreement across taxonomic hierarchies. However, taxonomic classifications of OTUs not associated with the human body, especially those from anaerobic environments such as non-human animal guts, soil samples or anaerobic digesters, may gain significant benefits from using a larger, more diverse training set, such as GG99.
Based on our results, we recommend that researchers implement larger, more diverse classification training sets, such as a 97–99% ID clustering of the GG database, in pipelines for processing HTP bacterial 16S rRNA gene sequencing surveys. The higher diversity of reference sequences had a significant impact on the amount of information provided by naïve Bayesian classification, including both higher abundance of classified reads and greater classification depth for each OTU. Another source for a large, diverse training set, which we did not test in this study, is the SSU SILVA 104 NR database available online (http://www.arb-silva.de; Pruesse et al., 2007). We expect this data set also to yield satisfactory results, based on its high diversity (>245 000 unique bacterial sequences). We also recommend that researchers trim reference sequences to reduce noise and represent only the primer-targeted region of the query data, and refrain from removing unclassified OTUs before measuring β-diversity.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Binladen J, Gilbert MT, Bollback JP Panitz F, Bendixen C, Nielsen R et al. (2007). The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One 2: e197.
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R . (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucl Acids Res 37 (Database issue): D141–D145.
Costello EK, Gordon JI, Secor SM, Knight R . (2010). Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J 4: 1375–1385.
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R . (2009). Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
DeSantis TZ, Keller K, Karaoz U, Alekseyenko AV, Singh NNS, Brodie EL et al. (2011). Simrank: rapid and sensitive general-purpose k-mer search tool. BMC Ecology 11: 11.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Felsenstein J . (1989). PHYLIP -- phylogeny inference package (version 3.2). Cladistics 5: 164–166.
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504.
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R . (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237.
Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P et al. (2007). DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucl Acids Res 35: e91.
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100.
Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML . (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PloS Genetics 4: e1000255.
Lauber CL, Hamady M, Knight R, Fierer N . (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120.
Liu ZZ, DeSantis TZ, Andersen GL, Knight R . (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucl Acids Res 36: e120.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z et al. (2008). The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog 4: e20.
Nawrocki EP, Kolbe DL, Eddy SR . (2009). Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337.
Neefs JM, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R . (1993). Compilation of small ribosomal subunit RNA structures. Nucl Acids Res 21: 3025–3049.
Price MN, Dehal PS, Arkin AP . (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PloS One 5: e9490.
Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked andaligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res 35: 7188–7196.
Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J et al. (2011). Responses of gut microbiota to weight loss in obese and lean mice. Obesity; ; e-pub ahead of print 19 May 2011.
Reeder J, Knight R . (2010). Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7: 668–669.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K et al. (2011). Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci USA 108: 4158–4163.
Acknowledgements
This study was supported by Grant UH2/UH3CA140233 from the Human Microbiome Project of the NIH Roadmap Initiative, the National Cancer Institute, NIH common fund contract U01-HG004866 (a Data Analysis and Coordination Center for the Human Microbiome Project), The Hartwell Foundation, the Arnold and Mabel Beckman Foundation, the David and Lucile Packard Foundation, Cornell University Agricultural Experiment Station federal formula funds NYC-123444 received from the USDA National Institutes of Food and Agriculture (NIFA), and USDA NIFA Grant 2007-35504-05381.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Werner, J., Koren, O., Hugenholtz, P. et al. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J 6, 94–103 (2012). https://doi.org/10.1038/ismej.2011.82
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2011.82
Keywords
This article is cited by
-
A tripartite bacterial-fungal-plant symbiosis in the mycorrhiza-shaped microbiome drives plant growth and mycorrhization
Microbiome (2024)
-
Intestinal microbiota composition of children with glycogen storage Type I patients
European Journal of Clinical Nutrition (2024)
-
Intensive antibiotic treatment of sows with parenteral crystalline ceftiofur and tulathromycin alters the composition of the nasal microbiota of their offspring
Veterinary Research (2023)
-
Effects of synbiotic supplementation on intestinal microbiota composition in children and adolescents with exogenous obesity: (Probesity-2 trial)
Gut Pathogens (2023)
-
Rapid and accurate taxonomic classification of cpn60 amplicon sequence variants
ISME Communications (2023)