Abstract
A common strategy of pathogenic bacteria is to form close associations with parasitic insects that feed on animals and to use these insects as vectors for their own transmission. Pathogens interact closely with other coexisting bacteria within the insect, and interactions between co-occurring bacteria may influence the vector competency of the parasite. Interactions between particular lineages can be explored through measures of α-diversity. Furthermore, general patterns of bacterial community assembly can be explored through measures of β-diversity. Here, we use pyrosequencing (n=115 924 16S rRNA gene sequences) to describe the bacterial communities of 230 prairie dog fleas sampled across space and time. We use these communinty characterizations to assess interactions between dominant community members and to explore general patterns of bacterial community assembly in fleas. An analysis of co-occurrence patterns suggests non-neutral negative interactions between dominant community members (P<0.001). Furthermore, bacterial communities of fleas shift dramatically across years (phylotype-based: R=0.829, P<0.001; phylogenetic-based: R=0.612–0.753, P<0.001), but they also significantly differ across space (phylotype-based: R=0.418, P<0.001; phylogenetic-based: R=0.290–0.328, P<0.001) and between flea species (phylotype-based: R=0.160, P=0.011; phylogenetic-based: not significant). Collectively, our results show that flea-associated bacterial communities are not random assemblages; rather, an individual flea's bacterial community is governed by interactions between bacterial lineages and by the flea's place in space and time.
Similar content being viewed by others
Introduction
Pathogenic bacteria pose serious threats to the health of humans, wildlife populations, and agricultural crops. In many cases, the spread of disease is mediated through arthropod vectors, and vector competency is a key parameter in epidemiological models. However, there is wide variation in vector competency between species (Eisen et al., 2006, 2007a, 2007b; Wilder et al., 2008b) and even between individuals of the same species (Wilder et al., 2008a). The underlying causes of these differences are clearly important, and a more thorough understanding of the ecological factors associated with the prevalence and persistence of pathogenic lineages associated with vectors will ultimately help predict and prevent the spread of disease.
When one animal parasitizes another, the host is exposed to many of the parasite's bacterial community members. As such, parasites are the primary vectors of many disease-causing bacteria. Ticks transmit the causative agent of Lyme Disease, Borrelia burgdorferi (Burgdorfer et al., 1985; Piesman et al., 1987; Parola and Didier, 2001), and Rickettsial diseases such as Rocky Mountain Spotted Fever (Dumler and Bakken, 1998; Parola and Didier, 2001; Parola et al., 2005); fleas transmit the causative agent of bubonic plague, Yersinia pestis (Perry and Fetherston, 1997; Gage and Kosoy, 2005), Rickettsial diseases such as Rickettsia felis (Adams et al., 1990; Azad et al., 1997; Raoult et al., 2001), and a number of diseases caused by strains of Bartonella (Chomel et al., 1996; Higgins et al., 1996). However, disease agents coexist with other non-pathogenic members of the diverse bacterial communities that all animals harbor. Entire bacterial communities of insects known to vector bacterial diseases have been described (for example ticks, fleas, and lice), but with very small sample sizes (Reed and Hafner, 2002; Benson et al., 2004; Jones et al., 2008). The bacterial communities of other blood-feeding invertebrates such as mosquitoes and leeches have also been described (Lindh et al., 2005; Worthen et al., 2006); somewhat surprisingly, however, these parasites are not known to vector pathogenic bacterial lineages. An unexplored factor that may affect the ability of pathogenic lineages to colonize and persist within an individual vector is the composition of the vector's bacterial community, whereby certain assemblages of bacteria or non-neutral interactions between specific lineages may alter the ability of specific pathogens to invade the community.
In addition to interactions with coexisting bacteria, pathogenic lineages also interact closely with their vectors and hosts. These interactions may affect the evolution of pathogenic lineages, especially if the pathogen relies on the vector or host for dispersal. Furthermore, some lineages are passed vertically from female vectors to their offspring (for example some lineages within the Rickettsiales and Bacteroidetes), and these lineages are particularly likely to coevolve tightly with the vector. Bacterial symbionts have been found to positively affect insect fitness (Haine, 2008; Pais et al., 2008). For example, vertically transmitted Wolbachia reduce the susceptibility of Drosophila to viral infections (Teixeira et al., 2008). Vertically transmitted bacteria may act similarly towards bacteria detrimental to insect health, and thereby affect the community assemblages.
Abiotic factors may also influence bacterial community assembly within disease vectors. Seasonal shifts in microbial communities have been observed in soil and marine communities (Pinhassi and Hagstrom, 2000; Riemann et al., 2000; Schadt et al., 2003; Lipson and Schmidt, 2004; Bjork et al., 2008), due in large part to changing environmental conditions. Similarly, epidemics of arthropod-vectored pathogenic lineages correlate with environmental parameters (Collinge et al., 2005; Augustine et al., 2008), suggesting that environmental conditions may affect the ability of at least some community members to persist.
To explore the factors affecting bacterial community assembly in vectors, we used pyrosequencing to describe the bacterial communities of prairie dog fleas sampled across space and time. From a community ecology perspective, this approach permits: (1) testing for non-neutral interactions between bacterial members within individual fleas through analyses of α-diversity and (2) testing for differences in bacterial communities across time, space, and flea species through analyses of β-diversity. Importantly, this is the first comprehensive survey of entire bacterial communities associated with a disease vector (in contrast to surveys aimed at detecting the prevalence of specific lineages) and the first to explore how time, space, or host-effects may correlate with the composition of those communities.
Materials and methods
Tissue collection
Black-tailed prairie dogs (n=39), Cynomys ludovicianus, were captured across Boulder County using live traps. Prairie dog fleas were sampled from six distinct colonies in 2004, and three colonies were sampled again in 2007; no obvious ecological differences exist between sites. Fleas were collected from prairie dogs from 23 February to 18 March 2004 and from 16 March to 17 April 2007. Sampling dates were chosen to maximize the potential of capturing Oropsylla tuberculata cynomuris (peak abundance in late winter and early spring), and dates differ between years due to variable weather conditions between years. For example, we only trapped if it was warm enough for prairie dogs to be active and if there was no snow on the ground; these conditions occurred later in 2007 than in 2004. Collected fleas were immediately placed in ethanol and stored at −20 °C until DNA extraction. Fleas collected in 2004 were identified based on morphology (Hubbard, 1947; Furman, 1982). Details of the sampling scheme (for example fleas per prairie dogs, prairie dogs per colony) are presented in Supplementary Table 1.
DNA extraction
Fleas were washed in a solution of 0.133 M NaCl, 1.11% sodium dodecyl sulfate, and 0.0088 M EDTA two times to remove as many bacteria from the external flea parts as possible. To maximize the removal of external bacteria, fleas were then subjected to a lysozyme (11.6 mg l−1) treatment for 30 min at 37 °C before being washed one final time. Cleaned fleas were crushed, and subjected to another lysozyme treatment of 30 min at 37 °C. After this final lysozyme treatment, DNA was extracted using the Qiagen DNeasy Tissue kit following the manufacturer's recommended protocol.
Molecular techniques
We sequenced the Cytochrome Oxidase II gene from fleas (n=184) to distinguish between Oropsylla hirsuta (n=140), Oropsylla tuberculata cynomuris (n=40), and a third unknown flea species (n=4) using the forward primer, COIIfLeu, and the reverse primer, COIIrLys (Maekawa et al., 1999). Template DNA, primers, and Qiagen hot-start PCR kit reagents were combined and subjected to the following conditions: 94 °C for 5 min; 35 cycles of 94 °C for 20 s, 55 °C for 30 s, and 72 °C for 50 s; 72 °C for 10 min. PCR products were cleaned by adding 1.5U Exonuclease I (New England Biolabs, Ipswich, MA, USA) and 0.75U Shrimp Alkaline Phosphatase (USB Corp., Cleveland, OH, USA) to each sample; 37 °C for 15 min followed by 80 °C for 15 min. Clean PCR products were sequenced by Functional BioSciences (Madison, WI, USA). Multiple haplotypes were detected in populations of O. hirsuta (n=16) and O. t. cynomuris (n=5); Accession #'s: FN546240–FN546260.
Bacterial communities of 230 fleas were described using bar-tagged 16S rRNA pyrosequencing primers (27F and 338R) that amplify variable region 2 (Hamady et al., 2008). Platinum PCR Supermix 96 kits (Invitrogen, Carlsbad, CA, USA) were used for all PCR reactions; this hot-start kit uses a high fidelity polymerase. To minimize any contamination of DNA in our PCR reaction mixes, we added 1/2U of Arctic Shrimp DNase and 1.5 μl of primers (6 mM solution of both primers) to each well in the kit and let sit at room temperature for 5–10 min. The PCR reaction mix (everything but template DNA) was then heated to 70 °C for 25 min to deactivate the DNase. Template DNA was added and PCR reactions occurred under the following conditions: 94 °C for 5 min; 40 cycles of 94 °C for 20 s, 52 °C for 45 s, 72 °C for 45 s; 72 °C for 10 min. PCR reactions were performed in triplicate for each DNA sample and negative controls were used. PCR products from all three reactions were combined and purified using Promega Wizard PCR Preps. The concentration of all PCR products (including negative controls) was determined using PicoGreen dsDNA reagent (Invitrogen) and samples were combined inverse to their concentration and prepared for pyrosequencing at the University of South Carolina's Environmental Genomics Core Facility.
Analyses
Bacteria from each flea were described using an oligonucleotide primer labeled with a unique barcode. Unique bar-tags permit the assignment of each individual DNA sequence to the correct individual flea subsequent to sequencing; details of this procedure are provided elsewhere (Hamady et al., 2008). Sequences were aligned using NAST (DeSantis et al., 2006) and then binned into phylotypes based on 97% sequence identity. Each sequence was classified according to the Ribosomal Database Project's taxonomy (http://rdp.cme.msu.edu/) and a consensus lineage was calculated for each phylotype. A few phylotypes (n=40) detected in negative controls were removed from all analyses. The contaminant DNA, all closely related β-proteobacteria, appears to have originated in the Invitrogen PCR kits; we are confident that sequences originating from contaminants were removed before analyses as they are not closely related to any of the commonly detected community members (Supplementary Figure 1).
Classifying phylotypes taxonomically provides an overall description of the bacterial communities associated with fleas, but the presence and absence of individual phylotypes within individual fleas can also be used to explore the interactions between bacterial lineages. To test for non-neutral interactions of bacterial lineages associated with fleas, we constructed a presence and absence matrix of any bacterial phylotype with an average relative abundance across all fleas of at least 0.4%. These 23 phylotypes (Table 1; Supplementary Figure 1) account for nearly 50% of all detected bacteria and interactions between these most common bacteria are most likely to be responsible for community dynamics. We used ECOSIM to test for non-neutral patterns of co-occurrence of bacterial phylotypes (Gotelli and Entsminger, 2004); row sums and column sums were held constant. ECOSIM tests for whether the observed distribution of phylotypes is significantly different from randomly assembled presence/absence matrices. As the algorithm used to generate phylotypes is heuristic and may split closely related lineages into separate phylotypes, especially with large datasets, we combined closely related lineages (1, 5, 14, and 23; 2 and 13; 4, 8, and 11; 7 and 9; 19 and 20; see Supplementary Figure 1) and repeated the ECOSIM analysis. We also partitioned the data by year and repeated these analyses.
Although α-diversity measurements compare the presence/absence patterns of specific lineages, β-diversity measurements compare one community to another using all community members in the comparison. The output of β-diversity measures is a pairwise distance matrix; in this case a representation of how similar each flea's bacterial community is to every other flea's bacterial community. This distance matrix can then be analyzed to see whether particular factors (in this case: time, space, and flea species) influence patterns of community assemblage. We used weighted and un-weighted UniFrac to measure phylogenetic β-diversity (Lozupone and Knight, 2005). For the phylotype-based analyses, phylotypes comprising at least 2% of the bacterial community in any one flea (n=240 phylotypes) were included in the analysis and abundances were normalized and fourth-root transformed (transformed so that highly abundant lineages do not dominate analyses). As a means to estimate β-diversity using the phylotype data, we transformed this modified phylotype abundance matrix with a Bray–Curtis transformation (transforms the ‘phylotype × sample’ abundance matrix to a ‘sample × sample’ distance matrix) using PRIMER-E (Lutton, UK). We used a two-way nested Analysis of Similarity to test for significant differences between groups (for example Year, Colony, and Flea species); null similarity distributions were calculated by repeatedly randomizing the input matrix (PRIMER-E). The overall contribution of individual phylotypes within a community to observed differences can be explored using SIMPER, an analysis tool within PRIMER-E (phylotype-based method only). The output of this analysis ranks each phylotype based on its percent contribution to differences.
Results and discussion
Bacterial diversity within individual fleas
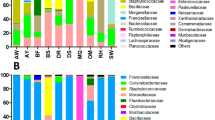
In all, 115 924 16S rRNA gene sequences were generated from 230 prairie dog fleas (number of sequences per flea; range=203–1300, mean=504, median=469, standard error=15). Sequences <200 basepairs in length were discarded and remaining sequences ranged from 200 to 296 basepairs in length (mean=228 basepairs). Sequences were assigned to 9986 unique phylotypes, but most of these phylotypes were rare. In all, 99.2% of detected phylotypes (9907/9986) had average relative abundances of <0.1%; similarly, 97.6% of detected phylotypes (9746/9986) comprised <2% of community membership in any individual flea. The 23 most abundant phylotypes represented nearly 50% of community membership (Table 1). Furthermore, 22 of the top 23 phylotypes were classified as Proteobacteria or Bacteroidetes (Table 1; Supplementary Figure 1). Also, only four phyla (Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes) represent >98%, on average, of individual bacterial communities in fleas (Figure 1). The diversity contained within these phyla is enormous, but the lack of diversity at the phylum-level corroborates previous suggestions that blood-feeding insects harbor bacterial communities low in diversity due to the immune response of host blood or low nutrient quality of blood (Graf et al., 2006; Worthen et al., 2006).
Phylum-level diversity and abundance in prairie dog fleas across (a) years, (b) flea species, (c) colonies in 2004, and (d) colonies in 2007. Average relative abundance within individual fleas: Proteobacteria (52.9%); Bacteroidetes (21.5%); Firmicutes (13.1%); Actinobacteria (10.1%); Unclassified (1.1%); Tenericutes (0.5%); Acidobacteria (0.2%); TM7 (0.2%); OD1 (0.1%); all other detected bacteria (<0.1%).
Measurements of α-diversity provide a means to explore interactions between lineages within a community. Here, each individual flea provides an estimate of community composition, and investigating a large number of these estimates (n=230) can lend insight into the types of interactions bacterial lineages have with one another (for example negative, neutral, or positive). For example, the presence of Rickettsia felis has been suggested to reduce overall bacterial richness (as estimated using RFLP data) within cat fleas, Ctenocephalides felis, presumably through negative interactions (Pornwiroon et al., 2007). Similarly, we find that bacterial lineages assemble non-randomly in fleas (P<0.001). We find the same non-random pattern when fleas are partitioned based on year (P<0.001), suggesting that our results are not due to stochastic variation of phylotype prevalence occurring between years. We also find the same patterns when closely related phylotypes are combined (P<0.001).
Interactions between bacterial lineages within arthropods have been suggested to alter population dynamics and virulence of pathogens (Lipsitch et al., 1996; Macaluso et al., 2002; Lively et al., 2005), but there have been few large-scale studies of bacterial communities in vectors to explore interactions between lineages. A recent study, however, explored community dynamics of five bacterial lineages (three were pathogenic lineages) within a large number of ticks (n=900) and found only neutral interactions between community members (Clay et al., 2008). However, the ability of this study to detect non-neutral interactions was limited by focusing on only five lineages. As we described flea-associated bacterial communities using universal bacterial primers, our results provide a more accurate estimate of bacterial community dynamics between all community members. Our study, however, is limited because we do not use a targeted detection approach; some lineages present in very low abundance may have gone undetected. Ultimately, combining the targeted detection approach with the universal detection approach may best improve our understanding of bacterial community interactions within disease vectors.
Bacterial diversity across individual fleas
β-Diversity measurements provide a means to test for patterns in bacterial community assembly (Lozupone et al., 2007). These measures have suggested that community assemblages correlate with salinity (Lozupone and Knight, 2007), diet (Ley et al., 2008), pH (Jones et al., 2009; Lauber et al., 2009), and host genotype (Khachatryan et al., 2008). We used a similar approach to explore the factors affecting bacterial community assembly in prairie dog fleas sampled across space and time. This is the first time that bacterial communities of a disease vector have been sampled at this depth or breadth. Weighted phylotype, weighted phylogenetic, and unweighted phylogenetic measures of β-diversity were used to assess patterns in bacterial community assemblages of fleas sampled across time, space, and flea species.
In prairie dog fleas of Boulder County, time (2004 vs 2007) had the greatest significant correlation with bacterial community assembly (R=0.612–0.829; Figure 2; Table 2). This suggests that shifts in communities occur range-wide across a large temporal scale. Bartonella and Rickettsiales are among the lineages responsible for differences across years; Bartonella decreased from 2004 to 2007, whereas Rickettsial abundance increased (Table 3). This result is consistent with the finding that Bartonella infection rates vary significantly across years in Boulder County prairie dogs (Bai et al., 2008). Notably, a number of lineages were detected in 2004, but were not detected or were very rare in 2007 (Table 3). Longitudinal studies of microbial communities have shown that soil and marine microbial communities shift through time (Pinhassi and Hagstrom, 2000; Riemann et al., 2000; Schadt et al., 2003; Lipson and Schmidt, 2004; Bjork et al., 2008), and changes in community assembly were attributed to shifts in environmental variables. In adult fleas, nutrient input consists of blood and changes in blood quality are unlikely to explain the inter-annual shift in communities. However, environmental factors (untested here) may affect the composition of the regional species pool that seeds flea-associated bacterial communities. A longitudinal study of vector-associated microbial communities in conjunction with the monitoring of environmental parameters would lend insight into this possibility.
3D PCoA plots of individual flea bacterial communities as determined using UniFrac (Lozupone and Knight, 2005). (3D PCoA plots were introduced in Ley et al., 2008). Communities are displayed according to (a) year (red: 2004; blue: 2007), and (b) flea species (red: O. hirsuta; blue: O. t. cynomuris; yellow: unknown flea species). Analyses of similarity between time, colony, and flea species are presented in Table 2.
In addition to temporal shifts in bacterial community composition, significant differences are also found across different prairie dog colonies within years (R=0.290–0.418; Table 2). Importantly, the phylotypes most responsible for differences among colonies include Bartonella spp. and lineages of Rickettsiales. This finding is consistent with a previous finding that Bartonella infection in Boulder County prairie dogs varies widely across colonies (Bai et al., 2008). Finally, almost all phylotypes occur in all colonies and differences between colonies are largely based on different abundance patterns of specific lineages, suggesting that dispersal constraints of certain lineages are not largely affecting community assembly and that differences between colonies are largely due to varied abundances of community members. It is unclear why there are significant differences among colonies. One possibility is that dispersal of community members is limited by the movement of fleas, which are in turn limited by the movement of their prairie dog hosts. Tests of genetic structure among fleas, using mtDNA sequences, and prairie dogs, based on microsatellite genotypes, revealed significant, but relatively weak, genetic structure for both taxa (data not presented). Alternatively, the degree of microbial community differentiation may reflect stochastic variation in the demography of the various bacterial species within the flea.
Among O. hirsuta and O. t. cynomuris, bacterial communities differ very slightly. The phylotype-based approach indicates that bacterial communities differ between flea species (R=0.160, P=0.011), but both weighted and unweighted UniFrac results indicate no differences between flea species (Table 2). Furthermore, correlations between community assemblages and flea species are much less than for year or colony. Despite the overall similarity of communities in the two flea species (the third unknown flea species was omitted from data analyses because only four were detected), there are notable differences in the abundances of individual lineages. Rickettsiales and Bartonella were again among the lineages most responsible for differences between flea species (phylotype-based method; Table 4). Rickettsiales and Bartonella were much less abundant in O. t. cynomuris than in O. hirsuta. Also, bacterial communities of O. hirsuta are much more variable than O. t. cynomuris communities (Figure 2). These two flea species differ widely in their ability to transmit Yersinia pestis, the causative agent of plague: O. t. cynomuris has more than three times greater transmission efficiency of bubonic plague than O. hirsuta (Wilder et al., 2008b). Perhaps the presence or absence of a specific lineage, such as the Rickettsial lineage studied here, or the difference in community composition variability across flea species accounts for the difference in vector competency. Unfortunately, Y. pestis was not detected in this study and the impact of community assembly on plague dynamics could not be analyzed.
Conclusions
Microbial communities in fleas differ substantially across time and space, but not between flea species or between individual prairie dogs. These results suggest that the characteristics of individual fleas or prairie dogs have relatively little effect on microbial communities within disease vectors. Instead, microbial communities may be strongly influenced by stochastic factors or currently unknown environmental factors that vary between years and among colonies. Without more samples across time, it is not possible to test the effects of prevailing climatic conditions or biotic factors. Similarly, too few colonies were surveyed to permit tests of whether particular factors that vary among colonies can explain some of the variation of community composition across space. However, the present results clearly point the way to an improved sampling scheme for future studies, and underscore the importance of spatial and time-series data in microbial ecology studies. Collectively, our results suggest that flea-associated microbial communities are not random assemblages of bacterial species present in the regional pool of fleas. The strong difference between communities from fleas collected in 2004 and 2007 indicates that major, region-wide shifts in flea-associated bacteria occur across time. Within years, however, significant differences in bacterial communities across prairie dog colonies are notable. Together, the temporal and spatial effects on bacterial community assembly in fleas parallel the temporal and spatial variation observed in Y. pestis and B. washoensis infection of prairie dogs (Collinge et al., 2005; Augustine et al., 2008; Bai et al., 2008). The overall lack of differentiation of bacterial communities between flea species suggests that O. hirsuta and O. t. cynomuris do not represent unique niches. However, certain lineages (for example Rickettsiales and Bartonella) differ substantially between the two flea species, and specific lineages may facilitate or inhibit pathogenic lineages within an individual flea.
Conflict of interest
The authors declare no conflict of interest.
References
Adams JR, Schmidtmann ET, Azad AF . (1990). Infection of colonized cat fleas, Ctenocephalides-Felis (Bouche), with a rickettsia-like microorganism. Am J Trop Med Hyg 43: 400–409.
Augustine DJ, Matchett MR, Toombs TP, Cully JF, Johnson TL, Sidle JG . (2008). Spatiotemporal dynamics of black-tailed prairie dog colonies affected by plague. Landsc Ecol 23: 255–267.
Azad AF, Radulovic S, Higgins JA, Noden BH, Troyer JM . (1997). Flea-borne rickettsioses: ecologic considerations. Emerging Infect Dis 3: 319–327.
Bai Y, Kosoy MY, Ray C, Brinkerhoff RJ, Collinge SK . (2008). Temporal and spatial patterns of Bartonella infection in black-tailed prairie dogs (Cynomys ludovicianus). Microb Ecol 56: 373–382.
Benson MJ, Gawronski JD, Eveleigh DE, Benson DR . (2004). Intracellular symbionts and other bacteria associated with deer ticks (Ixodes scapularis) from Nantucket and Wellfleet, Cape Cod, Massachusetts. Appl Environ Microbiol 70: 616–620.
Bjork RG, Bjorkman MP, Andersson MX, Klemedtsson L . (2008). Temporal variation in soil microbial communities in Alpine tundra. Soil Biol Biochem 40: 266–268.
Burgdorfer W, Lane RS, Barbour AG, Gresbrink RA, Anderson JR . (1985). The western black-legged tick, ixodes-pacificus—a vector of borrelia-burgdorferi. Am J Trop Med Hyg 34: 925–930.
Chomel BB, Kasten RW, FloydHawkins K, Chi BH, Yamamoto K, RobertsWilson J et al. (1996). Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol 34: 1952–1956.
Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C . (2008). Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol Ecol 17: 4371–4381.
Collinge SK, Johnson WC, Ray C, Matchett R, Grensten J, Cully JF et al. (2005). Landscape structure and plague occurrence in black-tailed prairie dogs on grasslands of the western USA. Landsc Ecol 20: 941–955.
DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al. (2006). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34: W394–W399.
Dumler JS, Bakken JS . (1998). Human ehrlichioses: newly recognized infections transmitted by ticks. Ann Rev Med 49: 201–213.
Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL . (2006). Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA 103: 15380–15385.
Eisen RJ, Lowell JL, Montenieri JA, Bearden SW, Gage KL . (2007a). Temporal dynamics of early-phase transmission of Yersinia pestis by unblocked fleas: secondary infectious feeds prolong efficient transmission by Oropsylla montana (Siphonaptera: Ceratophyllidae). J Med Entomol 44: 672–677.
Eisen RJ, Wilder AP, Bearden SW, Montenieri JA, Gage KL . (2007b). Early-phase transmission of Yersinia pestis by unblocked Xenopsylla cheopis (Siphonaptera: Pulicidae) is as efficient as transmission by blocked fleas. J Med Entomol 44: 678–682.
Furman DE, Catts EP . (1982). Order Siphonoptera. In: Manual of Medical Entomology. Cambridge University Press: Cambridge, p 207.
Gage KL, Kosoy MY . (2005). Natural history of plague: perspectives from more than a century of research. Ann Rev Entomol 50: 505–528.
Gotelli NJ, Entsminger GL . (2009). EcoSim: Null Models Software for Ecology. Acquired Intelligence Inc. & Kesey-Bear, Jericho, VT (http://www.garyentsminger.com/ecosim/index.htm).
Graf J, Kikuchi Y, Rio RVM . (2006). Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol 14: 365–371.
Haine ER . (2008). Symbiont-mediated protection. Proc R Soc B-Biol Sci 275: 353–361.
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R . (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237.
Higgins JA, Radulovic S, Jaworski DC, Azad AF . (1996). Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae). J Med Entomol 33: 490–495.
Hubbard CA . (1947). Fleas of Western North America. Hafner Publishing Company: New York.
Jones RT, McCormick KF, Martin AP . (2008). Bacterial communities of Bartonella-positive fleas: diversity and community assembly patterns. Appl Environ Microbiol 74: 1667–1670.
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N . (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3: 442–453.
Khachatryan ZA, Ktoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI . (2008). Predominant role of host genetics in controlling the composition of gut microbiota. PLOS ONE 3: e3064.
Lauber CL, Hamady M, Knight R, Fierer N . (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120.
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI . (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788.
Lindh JM, Terenius O, Faye I . (2005). 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol 71: 7217–7223.
Lipsitch M, Siller S, Nowak MA . (1996). The evolution of virulence in pathogens with vertical and horizontal transmission. Evolution 50: 1729–1741.
Lipson DA, Schmidt SK . (2004). Seasonal changes in an alpine soil bacterial community in the Colorado Rocky mountains. Appl Environ Microbiol 70: 2867–2879.
Lively CM, Clay K, Wade MJ, Fuqua C . (2005). Competitive co-existence of vertically and horizontally transmitted parasites. Evol Ecol Res 7: 1183–1190.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Lozupone CA, Hamady M, Kelley ST, Knight R . (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73: 1576–1585.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440.
Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF . (2002). Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J Med Entomol 39: 809–813.
Maekawa K, Kitade O, Matsumoto T . (1999). Molecular phylogeny of orthopteroid insects based on the mitochondrial cytochrome oxidase II gene. Zool Sci 16: 175–184.
Pais R, Lohs C, Wu YN, Wang JW, Aksoy S . (2008). The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol 74: 5965–5974.
Parola P, Didier R . (2001). Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis 32: 897–928.
Parola P, Paddock CD, Raoult D . (2005). Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev 18: 719.
Perry RD, Fetherston JD . (1997). Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev 10: 35.
Piesman J, Mather TN, Sinsky RJ, Spielman A . (1987). Duration of tick attachment and Borrelia-Burgdorferi transmission. J Clin Microbiol 25: 557–558.
Pinhassi J, Hagstrom A . (2000). Seasonal succession in marine bacterioplankton. Aquat Microb Ecol 21: 245–256.
Pornwiroon W, Kearney MT, Husseneder C, Foil LD, Macaluso KR . (2007). Comparative microbiota of Rickettsia felis-uninfected and—infected colonized cat fleas, Ctenocephalides felis. ISME J 1: 394–402.
Raoult D, La Scola B, Enea M, Fournier PE, Roux V, Fenollar F et al. (2001). A flea-associated rickettsia pathogenic for humans. Emerging Infect Dis 7: 73–81.
Reed DL, Hafner MS . (2002). Phylogenetic analysis of bacterial communities associated with ectoparasitic chewing lice of pocket gophers: a culture-independent approach. Microb Ecol 44: 78–93.
Riemann L, Steward GF, Azam F . (2000). Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol 66: 578–587.
Schadt CW, Martin AP, Lipson DA, Schmidt SK . (2003). Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301: 1359–1361.
Teixeira L, Ferreira A, Ashburner M . (2008). The bacterial symbiont wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. Plos Biol 6: 2753–2763.
Wilder AP, Eisen RJ, Bearden SW, Montenieri JA, Gage KL, Antolin MF . (2008a). Oropsylla hirsuta (Siphonaptera: Ceratophyllidae) can support plague epizootics in black-tailed prairie dogs (Cynomys ludovicianus) by early-phase transmission of Yersinia pestis. Vector Borne Zoonotic Dis 8: 359–367.
Wilder AP, Eisen RJ, Bearden SW, Montenieri JA, Tripp DW, Brinkerhoff RJ et al. (2008b). Transmission efficiency of two flea species (Oropsylla tuberculata cynomuris and Oropsylla hirsuta) involved in plague epizootics among prairie dogs. Ecohealth 5: 205–212.
Worthen PL, Gode CJ, Graf J . (2006). Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl Environ Microbiol 72: 4775–4781.
Acknowledgements
We thank all the individuals who graciously helped with prairie dog trapping, flea collection, and flea identification, especially Amelia Markeson, Katie McCormick, Adam Mitchell, and Jory Brinkerhoff. Thanks to Abimbola Kazeem for assisting with DNA sequencing of flea COII. Thanks to Micah Hamady for his help with data analyses (both development and implementation). We thank Boulder County Open Space and Boulder City Open Space and Mountain Parks for access to land and animals. Funding was provided by the following sources: Department of Ecology and Evolutionary Biology at the University of Colorado-Boulder; the William H Burt Fund of the University of Colorado Museum; the Beverly Sears Student Award program of the University of Colorado; the Colorado Mountain Club; a NSF Doctoral Dissertation Improvement Grant awarded to RTJ (DEB #0808560); and a NIH grant awarded to APM and RK (# R01 HG004872-01).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Rights and permissions
About this article
Cite this article
Jones, R., Knight, R. & Martin, A. Bacterial communities of disease vectors sampled across time, space, and species. ISME J 4, 223–231 (2010). https://doi.org/10.1038/ismej.2009.111
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2009.111
Keywords
This article is cited by
-
Laboratory colonization stabilizes the naturally dynamic microbiome composition of field collected Dermacentor andersoni ticks
Microbiome (2017)
-
Assessment of bacterial diversity in Hyalomma aegyptium, H. marginatum and H. excavatum ticks through tag-encoded pyrosequencing
Experimental and Applied Acarology (2017)
-
The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility
The ISME Journal (2016)
-
Path analyses of cross-sectional and longitudinal data suggest that variability in natural communities of blood-associated parasites is derived from host characteristics and not interspecific interactions
Parasites & Vectors (2015)
-
Similarities and seasonal variations in bacterial communities from the blood of rodents and from their flea vectors
The ISME Journal (2015)