Abstract
Purpose:
Blepharocheilodontic (BCD) syndrome is a rare autosomal dominant condition characterized by eyelid malformations, cleft lip/palate, and ectodermal dysplasia. The molecular basis of BCD syndrome remains unknown.
Methods:
We recruited 11 patients from 8 families and performed exome sequencing for 5 families with de novo BCD syndrome cases and targeted Sanger sequencing in the 3 remaining families.
Results:
We identified five CDH1 heterozygous missense mutations and three CTNND1 heterozygous truncating mutations leading to loss-of-function or haploinsufficiency. Establishment of detailed genotype–phenotype correlations was not possible because of the size of the cohort; however, the phenotype seems to appear more severe in case of CDH1 mutations. Functional analysis of CDH1 mutations confirmed their deleterious impact and suggested accelerated E-cadherin degradation.
Conclusion:
Mutations in CDH1 encoding the E-cadherin were previously reported in hereditary diffuse gastric cancer as well as in nonsyndromic cleft lip/palate. Mutations in CTNND1 have never been reported before. The encoded protein, p120ctn, prevents E-cadherin endocytosis and stabilizes its localization at the cell surface. Conditional deletion of Cdh1 and Ctnnd1 in various animal models induces features reminiscent of BCD syndrome and underlines critical role of the E-cadherin-p120ctn interaction in eyelid, craniofacial, and tooth development. Our data assert BCD syndrome as a CDH1 pathway–related disorder due to mutations in CDH1 and CTNND1 and widen the phenotypic spectrum of E-cadherin anomalies.
Genet Med advance online publication 09 March 2017
Similar content being viewed by others
Introduction
Blepharocheilodontic (BCD) syndrome (OMIM 119580) is a rare disorder characterized by eyelid malformations, cleft lip/palate (CLP), and dental anomalies.1 To date, 35 patients have been reported.2 CLP, which is usually bilateral, is the major feature. Eyelid malformations (ectropion of the lower eyelids, euryblepharon, and lagophthalmia) are typical. Patients have variable expressions of ectodermal dysplasia, with constant dental anomalies (i.e., conical teeth and tooth agenesis). In addition, hypothyroidism due to thyroid gland hypoplasia or aplasia, imperforate anus, neural tube defect, and syndactyly have been reported in a few patients. BCD syndrome is usually sporadic, but large affected families have been described, supporting autosomal dominant inheritance.1,2,3,4,5,6 Focusing on genes involved in palate and thyroid development and clinical similarities with conditions comprising ectodermal defects and oral clefts, preliminary molecular studies have failed to identify causative mutation in TP63, IRF6, TBX10, FOXE1, and OSR2 genes.7 The molecular basis of the condition thus remained unknown.
We recruited 11 patients from 8 families and performed exome sequencing of parent–child trio in 5 families with de novo BCD syndrome. The remaining families were subsequently studied via targeted Sanger sequencing. We identified five heterozygous missense mutations in CDH1 and three heterozygous truncating mutations in CTNND1. CDH1 encodes E-cadherin, a classical cadherin playing a central role in strong intercellular adhesion in epithelial cells.8 The protein is highly expressed in human and mice embryos during critical stages of lip and palate development.9,10 Several mouse models highlighted critical roles of CDH1 for eyelid, craniofacial, tooth, and hair development.11,12,13,14 Mutations in CDH1 cause early-onset, multigenerational hereditary diffuse gastric cancer and lobular breast cancer (HDGC; OMIM 137215).15,16 At least six families have been reported with segregating HDGC and CLP,9,17,18 and recently mutations in CDH1 were also reported in nonsyndromic CLP (NSCLP).19,20 CTNND1 encodes the catenin delta-1 (alias p120ctn), an Armadillo repeat protein that interacts closely with the E-cadherin. The intracellular domain of E-cadherin binds to p120ctn, stabilizing the E-cadherin complex at the membrane and preventing its binding to the endocytosis machinery.21 CTNND1 has not previously been reported as a disease-causing gene. In Xenopus laevis, the gene is highly expressed in the craniofacial skeleton and epidermidis.22 Interaction between E-cadherin and p120ctn plays a major role in eyelid and tooth development in mice.9,11,12,13,14 We performed functional studies to ascertain the deleterious consequences of the CDH1 missense mutations identified in BCD syndrome patients and concluded that CDH1 and CTNND1 are responsible for this condition.
Materials and Methods
Patients
Patients were recruited by a French national collaboration referred by the multidisciplinary cleft clinic of Lille University Hospital and the national centers for Rare Developmental Diseases. Only patients fulfilling diagnostic criteria were selected (eyelid anomalies, CLP or choanal atresia, and ectodermal dysplasia). Our cohort comprised 11 patients from 8 families. In 6 families, BCD syndrome occurred sporadically; in 2 families, the condition is transmitted with an autosomal dominant pattern.
Ethics statement
This study was conducted according to the Declaration of Helsinki and with written consent to use clinical data and photographs for research purposes obtained from the patients, parents, or legal representative.
Library preparation, exon capture, and high-throughput sequencing
DNA samples from participating individuals were sent to the Microarray and Sequencing platform of the Institut de Génétique et de Biologie Moléculaire et Cellulaire, a member of the France Génomique consortium (ANR-10-INBS-0009). Genomic DNA (1 µg) was sheared to obtain a mean fragment size of 250 nt using Covaris E210 (Covaris, Brighton, UK), followed by library preparation using a SureSelect XT2 Reagent kit (Agilent Technologies Courtaboeuf, france). Exon capture was performed on barcoded libraries pooled by eight using the SureSelect XT2 Human All Exon V5 enrichment System (Agilent Technologies) following the manufacturer’s protocol. DNA libraries were checked for quality and quantified using a 2100 Bioanalyzer (Agilent Technologies) and loaded at a concentration of 8 pM in the flow cell, multiplexed by four per lane, and sequenced using a Hiseq 2500 (Illumina, Eindhoven, The Netherlands) as paired-end 2 × 100 base reads following the manufacturer’s protocol.
Bioinformatics analysis
Image analysis and base calling were performed using the Illumina RTA (Real-Time Analysis) v1.18.61 software, and FASTQ files were generated and demultiplexed with CASAVA v1.8.2. Reads were aligned onto the hg19 assembly of the Homo sapiens genome using BWA v0.7.5a.23 Aligned data were refined with Picard v1.122 (http://picard.sourceforge.net/) to flag duplicate reads and GATK v3.2-224 to perform local realignments and to recalibrate base qualities. Samtools v0.1.1925 was used to filter out multimapped reads. Variant calling was performed using a GATK UnifiedGenotyper, and variant quality scores were recalibrated using the GATK VariantRecalibrator tool. Variants were annotated using the GATK VariantAnnotator, SnpEff v2.0.5, and SnpSift v3.3c. Finally, VaRank v1.2.426 was used to rank discovered variants.
Sanger sequencing
PCR amplification and Sanger sequencing of the complete coding regions of CDH1 (NM_004360.3) and CTNND1 (NM_001085458.1), including exon–intron boundaries and UTRs, were performed. Primers are available on request.
In silico mutation analyses
Protein changes due to CDH1 and CTNND1 mutations were predicted using Alamut2.7.2 (Interactive Biosoftware). The possible impact of amino acid substitutions on the structure and function of E-cadherin was predicted using the following bioinformatics tools: PolyPhen-2 (http:/genetics.bwh.harvard.edu/pph2/), Align GVGD (http://agvgd.iarc.fr/agvgd_input.php), SIFT Aligned Sequences (http://sift.jcvi.org/www/SIFT_aligned_seqs_submit.html), and Mutation Taster (http://www.mutationtaster.org/). Amino acid sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/)
The human native EC1 and EC2 domains of E-cadherin protein and the human Asp254Tyr and Asp257Val mutated EC1 and EC2 domains of E-cadherin proteins were predicted with the Phyre2 server (Protein Homology/analogY Recognition Engine V2.0) using the deduced amino acid sequence of each protein. The predicted 3D structure of proteins was compared with the 3D resolved structure of the human native EC1 and EC2 domains of E-cadherin protein in dimeric conformation (PDB code: 1EDH) (100% identity sequence with the human native E-cadherin protein and 99% with both human mutated E-cadherin proteins) using the molecular visualization system, RasMol 2.7.5.1 (Herbert J. Bernstein). The topology of proteins was conserved in all the 3D-predicted structures (Figure 2B, a).
Plasmid constructions
Construction containing human E-cadherin GFP was a gift from Jennifer Stow (Addgene plasmid 28009).27 Using the QuikChange XL directed-mutagenesis kit (Stratagene, La Jolla, CA), we generated mutant versions of the constructions introducing the five CDH1 mutations identified and the HDGC-associated mutation p.(Asp244Gly) used as positive control.
Cell culture and transfection
Human embryonic kidney HEK293T cells 5 (ATCC CRL-1573) were grown and maintained in DMEM medium supplemented with 10% fetal bovine serum , 0.5% penicillin streptomycin, and 1% l-glutamine. Cells were seeded at 30,000 cells per well in six-well culture plates 24 h before transfection. HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen, Fisher Scientific, Illkirch, France) for 24 h following the manufacturer’s instructions. Empty vector pcDNA3.1 or pcDNA3.1-CDH1-GFP wild type or mutants were individually transfected. The total amount of DNA transfected to each cell was kept constant at 1,000 ng. Cells tested negative for mycoplasma contamination.
Western blot
Equal amounts of protein were subjected to 4–12% SDS-PAGE. We used anti-E-cadherin antibody (clone HECD-1, ab1416; Abcam, Cambridge, UK), anti-p120ctn antibody (NBP1-85383, rabbit polyclonal; Novus Biological), and horseradish peroxidase conjugated secondary antibodies (ThermoFisher, Fisher Scientific, Illkirch, France). Western blotting and quantification were performed using LAS4000 (Fujifilm) and ImageJ software (https://imagej.nih.gov/ij/).
Immunofluorescent staining
Effects of CDH1 mutations on E-cadherin subcellular localization were studied by fluorescence microscopy. Immunofluorescent staining was performed following standard procedures. Cells were fixated in 4% paraformaldehyde for 20 min at room temperature. Blocking and antibody incubations were performed in phosphate-buffered saline with 0.2% Triton X-100 and 2% bovine serum albumin. Cells were incubated with a specific primary anti-E-cadherin antibody (clone HECD-1, ab1416; Abcam, Cambridge, UK) and anti-p120ctn antibody (NBP1-85383, Rabbit polyclonal; Novus Biological), followed by secondary antibodies Alexa Fluor 555 and 488 (Molecular Probes, Fisher Scientific). Fluorescence images of cells were captured and analyzed with an LSM 710 confocal microscope (Carl Zeiss, Jena, Germany).
Results
Clinical phenotype
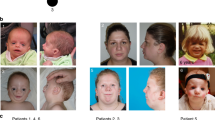
All patients involved in this study had typical BCD syndrome with variable expression severity ( Table 1 ). Eyelid anomalies were constant, including ectropion of the lower eyelids, euryblepharon (10/11), lagophthalmia (10/11), and distichiasis (10/12). Features of ectodermal dysplasia were found in all patients (hair anomalies, conical teeth, tooth agenesis). CLP was observed in 9 of 11 patients. We also noted congenital hypothyroidism (thyroid gland hypoplasia or agenesis) in four patients, anal atresia in two patients, syndactyly in two patients, and neural tube defects in two patients. Vertex aplasia was observed in only two patients, from the same family (family 4). Both patients from family 3 had allodynia. Furthermore, all patients had typical dysmorphism with hypertelorism, a flat face, and a high forehead; some had an asymmetrical face ( Figure 1 ). There was no familial history of gastric cancer in these families except in family 4, in which the mother of F4-I2 had been diagnosed with gastric cancer and died at the age 35 years. We had no further information about the tumor histopathology. However, we could observe from photographs that she was not dysmorphic, and her daughter said that she had no feature reminiscent of BCD syndrome.
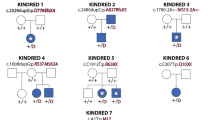
Pedigrees and clinical features of affected individuals with CDH1 and CTNND1 mutations. (A) Individuals for whom exome sequencing was performed are indicated by asterisks (*). +/- and -/- refer to heterozygous carriers of CDH1 and CTNND1 mutations and to nonmutation carriers, respectively. Pedigrees show autosomal dominant inheritance in families 4 and 8 and de novo mutation in patients F3-II1, F3-II2, and F7-II2. Individuals F1-I1 and F6-I1 are asymptomatic carriers of CHD1 and CTNND1 mutation. (B-a) Monozygous twins from family 4 and patient F4-II1 showing hair dysplasia, high domed forehead, and scar due to cleft lip/palate surgery. (B-b) Eyelid anomalies were more severe in patients with CDH1 mutations (F1-II1–F4-II2) compared to CTNND1 mutations (F7-II1–F8-II1), especially lower-eyelid ectropion and euryblepharon.
Identification of mutations in CDH1 and CTNND1
We performed whole-exome sequencing for five unrelated individuals in a parent–child trio strategy (families 1, 2, 3, 6, and 7), except for family 2 because DNA samples from individual F2-I1 were not available. We systematically selected candidate de novo events based on protein-altering and splice-site DNA changes absent from public databases (ClinVar https://www.ncbi.nlm.nih.gov/clinvar/, 1000 Genomes Project http://www.1000genomes.org/dbSNP/, ExAC http://exac.broadinstitute.org/) supported by at least two reads and 20% of total reads in the proband. This analysis identified 15 to 22 de novo variants per proband. After the alignment for each variant had been reviewed, the only candidates remaining on the basis of gene function and expression were one variant in CDH1 (NM_004360.3) c.1320G>T p.(?) in family 3 and one truncating variant in CTNND1 (NM_001085458.1) c.1093C>T p.(Gln365*) in family 7. Because the association was biologically plausible, we reanalyzed data from families 1, 2, and 6. We identified two additional variants in CDH1 (c.760G>T p.(Asp254Tyr) and c.770A>T p.(Asp257Val) and one truncating variant in CTNND1 (c.606_627del p.(Pro203Leufs*25)) in family 6, providing further arguments for the involvement of these genes in the condition.
Sequencing of the coding sequence and intron–exon boundaries of CDH1 and CTNND1 in four additional individuals from three families (families 4, 5, and 8) led to the identification of three additional mutations, c.1320 + 1G>C p.(?) and c.1361_1369del p.(Val454del) in CDH1 (families 4 and 5, respectively) and c.2098C>T p.(Arg700*) in CTNND1 (family 8). All affected residues were highly conserved and the substitutions were predicted to be deleterious by various in silico methods. In families 3 and 7, the mutations occurred de novo; in families 4 and 8, BCD syndrome cosegregated with the mutations. In families 1 and 6, the mutations were inherited from an asymptomatic parent. In families 2 and 5, parental DNA samples were not available, precluding the segregation study ( Figure 1A ). We did not find convincing variation in other genes, including TP63, IRF6, TBX10, FOXE1, CDH3, and genes involved in the E-cadherin signaling pathway.
Functional analysis of the mutations
We studied the functional effects of the CDH1 missense mutations. E-cadherin plays a central role in the strong intercellular adhesion in epithelial cells. The protein comprises an extracellular domain, a transmembrane domain, and an intracytoplasmic domain. The E-cadherin extracellular domain is characterized by five repeats of approximately 110 amino acids, each corresponding to a protein module of immunoglobulin-like fold called “extracellular cadherin” or “EC” domain (EC1 to EC5). The EC1 domain is folded into a seven-stranded beta-stand (A to G). Interconnections between successive domains are rigidified by conserved calcium ion–binding sites that support the rodlike conformation of the extracellular region.8 The connection between EC1 and EC2, also called “linkers,” contains the “calcium binding pocket,” i.e., the conserved Asp254-Gln255-Asn256-Asp257 sequence.28 Side chains of residues Asp254 and Asp257, negatively charged, directly interact with the calcium ions.29 Substitutions p.(Asp254Tyr) and p.(Asp257Val) possibly disrupt the electrostatic interactions between EC1–EC2 linker and calcium ions. In addition, a three-dimensional prediction of mutated E-cadherin shows changes in the orientation of the side chain of amino acids, particularly concerning the p.(Asp254Tyr) substitution, which probably impairs interactions with calcium ions ( Figure 2B , a ). Mutations c.1320G>T and c.1320 + 1G>C disrupt the canonical consensus donor motif AGgt, which is critical for splicing ( Figure 2B , b ). Both mutations are predicted to induce exon 9 skipping. RT-PCR on lymphocytes RNA of individual F5-I2 and sequencing confirmed exon 9 skipping due to c.1320G+1G>C mutation ( Figure 2B , c ). RNA samples from individual F4-II1 were not available. Exon 9 skipping is expected to induce removal of the major portion of the EC3 domain (from residue Tyr380 to residue Lys440), presumptively impairing its adhesive function. Finally, the p.(Val454del) deletion removes a conserved hydrophobic residue located at the C-terminal end of a beta-strand of the EC3 domain. Alignment of human EC1 and EC3 domains (data not shown) suggested that Val454 might correspond to residue Val235, which is critical for EC1–EC2 dimerization.30 Thus, residue Val454 may be involved in EC3 adhesive function. CTNND1 comprises 21 exons and encodes p120ctn, which is expressed as four isoforms that arise by alternative splicing from four start codons (M1 to M4). Isoforms 1 and 3 are the most widely and abundantly expressed isoforms. Exons 18 (exon A), exon 20 (exon B), and exon 11 (exon C) undergo alternative splicing.31 Mutations identified in CTNND1 are located, respectively, in exon 6 (c.606_627del p.(Pro203Leufs*25)), exon 7 (c.1093C>T p.(Gln365*)), and exon 14 (c.2098C>T p.Arg700*)) and are expected to affect isoforms 1 to 3. Truncating mutations in CTNND1 probably lead to nonsense-mediated RNA decay (NMD), an evolutionary conserved mRNA quality control system that degrades transcripts containing premature termination codons located more than 50–54 nucleotides upstream of the last exon–exon junction.32 Therefore, mutations identified in CTNND1 probably lead to haploinsufficiency. Less likely, truncating mutations lead to the production of a short protein lacking important functional domains ( Figure 2A , b ).
CDH1 mutations analysis. (A) (a) Structure of E-cadherin comprising protein signal peptide, precursor sequence, extracellular domain (EC), transmembrane domain (TM), and cytoplasmic domain. Exons and binding domain of p120ctn are indicated. E-cadherin extracellular domain is characterized by five repeats EC domains (EC1 to EC5). EC1–EC2 interconnection shows the conserved DQND sequence (surrounded) corresponding to the calcium ion–binding sites. Residues Asp254, Asp257, and Val454 show high conservation across vertebrates. Location of the missense germ-line E-cadherin mutations identified in BCD syndrome (indicated by asterisks), HDGC without or with cleft lip/palate (HDGC+CLP in blue), and NSCLP (in green). (A) (b) Structure of p120ctn. Four isoforms arise by alternative splicing from four start codons (M1 to M4). Isoforms 1 and 3 are the most abundantly expressed isoforms. Exons 18 (exon A), exon 20 (exon B), and exon 11 (exon C) undergo alternative splicing. CTNND1 mutations identified in our cohort, indicated by asterisks, affect isoforms 1 to 3. (B) (a) Three-dimensional predicted structure of the human native and mutated (Asp254Tyr and Asp257Val) E-cadherin proteins. EC1 and EC2 domains are colored orange, with residues 254 and 257 shown in CPK. Both substitutions possibly disrupt electrostatic interactions between calcium ions and mutated binding side. Substitution p.(Asp254Tyr) possibly further impairs interactions through modification of orientation of the side chain. (B) (b) Mutations c.1320G>T and c.1320G+1G>C disrupt the canonical consensus motif AGgt at the exon 9–intron 9 boundary. 3D structures are visualized with RasMol 2.7.5.1 (Herbert J. Bernstein). (c) RT-PCR on lymphocytes RNA from individual F5-I2 and healthy control. A shorter transcript was identified in patient F5-I2, corresponding to skipping of exon 9. Sequencing confirms exon skipping.
E-cadherin expression assay
Western blot revealed no detectable E-cadherin protein in HEK293T cells transfected with constructs expressing mutations c.760G>T p.(Asp254Tyr), c.1320G>T p.(?), and c.1320 + 1G>C p.(?). Compared to the wild type, cells expressing substitution c.1361_1363del p.(Val454del) showed decreased E-cadherin protein expression (−50%; P = 0.05), whereas cells expressing substitution c.770A>T p.(Asp257Val) showed only slightly decreased E-cadherin protein expression (−6%; P = 0.05). Interestingly, no quantitative expression of E-cadherin protein was observed in cells expressing the HDGC-associated substitution p.(Asp244Gly) compared to the wild type ( Figure 3b ).
Functional analysis of CDH1 mutations. (a) E-cadherin expression levels in cells expressing wild-type (wt) and mutated CDH1. The HDGC-associated mutation p.(Asp244Gly) is used as a positive control. There is no detectable E-cadherin protein for mutations p.(Asp254Tyr) and del exon 9 (c.1320G>T and c.1320 + 1G>C inducing exon 9 skipping). (b) Representation of the relative quantification of total E-cadherin. Compared to wild type, substitution p.(Val454del) shows a dramatic decrease in E-cadherin protein expression (−50%; P = 0.05), whereas substitution p.(Asp257Val) shows only a slight decrease (−6%; P = 0.05). Substitution p.(Asp244Gly) showed equal amount of E-cadherin protein compared to wild type. Significant P-values are indicated by * (P < 0.05). (c) Confocal microscopy images of HEK239T cells expressing wt and mutated CDH1 double-labeled with anti-E-cadherin antibody (green) and p120ctn antibody (red). HEK293T cells do not express detectable E-cadherin. In CDH1 wt-expressing cells, E-cadherin is mainly located at the plasma membrane, colocalizing with p120ctn. Mutations identified in CDH1 induce loss of cytoplasmic membrane staining and p120ctn colocalization and show intracytoplasmic perinuclear accumulation. The HDGC-associated mutation p.(Asp244Gly) also induced loss of cytoplasmic membrane localization and intracytoplasmic E-cadherin accumulation.
E-cadherin subcellular localization
In cells expressing wild-type CDH1, E-cadherin is mainly located at the plasma membrane and colocalize with p120ctn. The CDH1 mutations, identified in our patients, induce loss of cytoplasmic membrane staining and intracytoplasmic perinuclear E-cadherin accumulation. The HDGC-associated mutation p.(Asp244Gly), used as control, also induced loss of cytoplasmic membrane localization and intracytoplasmic E-cadherin accumulation ( Figure 3c ). Taken together, the in vitro results indicate that the variants affect E-cadherin expression and its subcellular localization and thus can be considered likely pathogenic mutations.
Discussion
Phenotypic description
We report 11 further patients presenting with typical BCD syndrome, thereby enhancing its delineation. All patients in our cohort had eyelid anomalies and ectodermal dysplasia, which are cardinal features. They also exhibited typical dysmorphism consisting of hypertelorism, a flat face, a high forehead, and, less frequently, an asymmetrical face ( Table 1 ; Figure 1B ). We have provided further evidence for the occurrence of congenital hypothyroidism, anal atresia, syndactyly, and neural tube defect in this condition. Vertex aplasia and allodynia, which were present, respectively, in family 4 and family 3, have never been reported before. CLP, thought to be constant, was absent in F1-II1 and F8-I2, whereas patient F1-II1 presented with unilateral choanal atresia. Finally, patient F8-I2 had a striking mild phenotype with only ectropion of lower eyelids and ectodermal dysplasia, including dental anomalies. The diagnosis was made retrospectively, when her son (F8-II2) was diagnosed with typical BCD syndrome ( Figure 1A ; Table 1 ). Therefore, our data suggest not only great interindividual but also intrafamilial variability, consistent with Weaver and colleagues’ description of a large BCD-syndrome family.6 Recently, a young girl harboring a syndromic CLP and a CDH1 mutation was reported.33 She had choanal atresia and a neural tube defect (meningoencephalocele) associated with developmental delay (probably due to the neural tube defect) and cardiac malformation (tetralogy of Fallot). The photograph provided in the article shows eyelid anomalies consistent with ectropion and euryblepharon, making it possible to retrospectively diagnose BCD syndrome.
CDH1 and CTNND1 mutations in BCD syndrome
Using exome sequencing for five families, and later direct sequencing of CDH1 and CTNND1 for three additional families, we identified either CDH1 (5/8) or CTNND1 (3/8) heterozygous mutations in all the families studied. Furthermore, the literature review drew our attention to a patient described by Nishi et al.,33 who carried a de novo CDH1 missense mutation c.2028C>A p.(Asp676Glu) and features reminiscent of BCD syndrome, possibly expanding this series to nine families harboring BCD and a CDH1-pathway anomaly. BCD is an autosomal dominant disorder with expression variability2-54 and is most often due to neomutation, or being inherited from an affected parent. Our findings are consistent with the following knowledge: when the familial study was available (6/8), most of the identified mutations either occurred de novo (2/6: CDH1 c.1320G>T p.(?) and CTNND1 c.1093C>T p.(Gnl365*)) or were inherited from an affected parent (2/6: CDH1 c.1320 + 1G>C and CTNND1 c.2098C>T) with wide expression variability. However, both CDH1 c.760G>T p.(Asp254Tyr) and CTNND1 c.606_627del p.(Pro203Leufs*25) were inherited from an a priori asymptomatic parent, suggesting incomplete penetrance of CDH1 and CTNND1 mutations in BCD syndrome. Although our series is too small to establish genotype–phenotype correlations, we observed that eyelid anomalies were more severe in patients with CDH1 mutations than in those with CTNND1 ( Figure 1B ). Furthermore, the mother of patient F4-I2, who had no features of BCD, had died of fulminant gastric cancer at age 35 years and was presumed to have carried the CDH1 mutation. Therefore, in accordance with recent reports involving CDH1 mutations in NSCLP,19,20 we can conclude that CDH1 mutations induce a wide spectrum of phenotypes defining CDH1-related disorders, ranging from asymptomatic carriers to NSCLP, HDGC with or without CLP, and BCD syndrome. Mutations identified in the various CDH1-related disorders have similar repartition along the E-cadherin protein and induce similar deleterious consequences9,15,19,20,34 ( Figure 2A , a ). Therefore, it is still unclear whether the different phenotypic expression of CDH1-related disorders reflects a genotype/phenotype correlation or an expansion of the phenotypic spectrum associated with CDH1 mutations.
CDH1-related disorders and preventive measures
Germ-line mutations in CDH1 cause HDGC. Carriers have a 70 to 80% lifetime risk of developing diffuse gastric cancer (also called signet ring cell gastric cancer) and lobular breast cancer.35,36 Owing to the high penetrance of these mutations, prophylactic total gastrectomy is currently recommended for CDH1 mutation carriers.16 However, to our knowledge, there are reports of at least one family presenting only with lobular breast cancer and two patients without International Gastric Cancer Linkage Consortium (IGCLC) criteria.37 Eight families have been described with NSCLP, among whom two carry CDH1 truncating mutations,18,19,20 but have no history of cancer. To date, in BCD syndrome, neither HDGC nor lobular breast cancer has ever been reported, even in large families.2,3,6 In our cohort, the five families with mutations in CDH1 did not meet IGCLC criteria. Gastric cancer history was noted only in the mother of F4-I2, without confirmation of diffuse gastric cancer. Therefore, one would have reservations about recommending an invasive procedure with high morbidity and mortality to a family with little or no history of cancer. As concluded in previous studies,17,37 CDH1 mutation penetrance in HDGC needs further assessment to better advise NSCLP, BCD syndrome, and asymptomatic families with pathogenic mutations regarding preventive measures.
Is BCD syndrome due to increased E-cadherin degradation?
The current landscape of CDH1 mutations associated with HDGC and NSCLP does not suggest preferential distribution of mutations along E-cadherin protein9,18,19,20 or genotype–phenotype correlation between truncating and missense mutations. However, it has previously been suggested that each CDH1 missense mutation probably has cell-specific biological behavior with distinct clinical impact,20,34 possibly supporting the CDH1-related disorders spectrum. Because all the mutations we identified in CDH1 were missense, we focused on this class of mutation. Ex vivo analyses of HDGC-associated missense mutations (listed in Figure 2A , a ) showed that abnormally folded proteins are retained within the endoplasmic reticulum, where they undergo ubiquitinylation and subsequent degradation through the endoplasmic reticulum–associated degradation (ERAD),38 leading to perinuclear accumulation and accelerated E-cadherin degradation. Mutations associated with NSCLP19,20 and BCD syndrome (this study) showed comparable deleterious effects. Nevertheless, decreased E-cadherin protein expression occurs to a greater degree in BCD syndrome, linking molecular anomalies induced by CDH1 missense mutations to CTNND1 haploinsufficiency. E-cadherin deprived of p120ctn interacts with other proteins, such as clathrin adapter proteins and Hakai, promoting E-cadherin endocytosis and degradation.39 Therefore, accelerated E-cadherin degradation may be a molecular basis underlying BCD syndrome. Regarding mutation p.(Asp257Val), the observed slightly decreased expression is probably due to escape from ERAD; therefore, other undetermined deleterious mechanisms may be involved in the occurrence of the phenotype.
E-cadherin and p120ctn are involved in craniofacial, eyelid, and tooth development
In human and mice embryos, CDH1 is highly expressed during critical stages of lip and palate development.9,10 In humans, E-cadherin is identified at 4 and 5 weeks in the frontonasal prominence and at 6 weeks in the lateral and medial nasal prominences. In X. laevis, p120ctn is highly expressed in the eye vesicles and the cranial neural crests, which are critical for cranial skeleton development.22 In mice, p120ctn is also highly expressed in teeth.11 Eyelid anomalies could be due to impairment of the Rock-I pathway. Indeed, p120 appears to recruit Rock-I to the E-cadherin complex and facilitate its activation locally. The protein complex p120/Rock-I promotes contractility in actin filaments, a critical process for embryonic eyelid closure.14 Rock-I knockout mice exhibited an “open eye at birth” phenotype, consistent with euryblepharon and lagophthalmia.13 Conditional targeting of E-cadherin in skin induces misshapen whiskers and sparse pelage hair reminiscent of the hair defect of BCD patients.40 Finally, conical teeth and teeth agenesis could be explained by the loss of E-cadherin and p120ctn interactions during ameloblast development. These columnar cells, originating from the labial cervical loop, are responsible for enamel development and are attached to the stratum intermedium at their basal end. In mice, deletion of E-cadherin disrupts the structure of the labial cervical loop and causes the separation of mature ameloblasts from the stratum intermedium.12 Protein p120ctn stabilizes ameloblast cadherins and maintains the tall and columnar organization of the ameloblasts at the secretory stage. Deletion of p120ctn does not disrupt tooth formation, but it prevents secretion of enamel matrix to initiate enamel mineralization.11,12 All these data support the hypothesis of CDH1 and CTNND1 involvement in BCD syndrome and disturbance of E-cadherin turnover as the possible molecular basis of the condition.
Conclusion
In 11 BCD patients from eight families, we identified five CDH1 deleterious missense mutations and three CTNND1 truncating mutations. A twelfth patient with CDH1 missense mutation has recently been reported with a phenotype reminiscent of BCD syndrome.33 Animal models conditionally deleting either Cdh1 or Ctnnd1 show striking similarities to patients, thereby supporting the involvement of both genes in the condition.11,12,22,40 BCD syndrome widens the phenotypic spectrum of E-cadherin anomalies and is thus a CDH1 pathway–related disorder that is probably due to accelerated E-cadherin degradation.
Disclosure
The authors declare no conflict of interest.
References
Gorlin RJ, Zellweger H, Curtis MW, et al. Blepharo-cheilo-dontic (BCD) syndrome. Am J Med Genet 1996;65:109–112.
Ababneh FK, Al-Swaid A, Elhag A, Youssef T, Alsaif S. Blepharo-cheilo-dontic (BCD) syndrome: expanding the phenotype, case report and review of literature. Am J Med Genet A 2014;164A:1525–1529.
Iida A, Narai S, Takagi R, Ono K, Ikeda N. Blepharo-cheilo-dontic (BCD) syndrome: case report. Cleft Palate Craniofac J 2006;43:237–243.
Gorlin RJ, Wiedemann HR. Blepharo-cheilo-dontic (BCD) syndrome. Acta Ophthalmol Scand Suppl 1996;219:22.
Guion-Almeida ML, Rodini ES, Kokitsu-Nakata NM, Bologna-Amantini D. Blepharo-Cheilo-Dontic (BCD) syndrome: report on four new patients. Am J Med Genet 1998;76:133–136.
Weaver KN, Rutledge KD, Grant JH, Robin NH. Imperforate anus is a rare associated finding in blepharocheilodontic syndrome. Am J Med Genet A 2010;152A:438–440.
Freitas EL, Martinhago CD, Ramos ES, Murray JC, Gil-da-Silva-Lopes VL. Preliminary molecular studies on blepharocheilodontic syndrome. Am J Med Genet A 2007;143A:2757–2759.
Patel SD, Chen CP, Bahna F, Honig B, Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr Opin Struct Biol 2003;13:690–698.
Frebourg T, Oliveira C, Hochain P, et al. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J Med Genet 2006;43:138–142.
Montenegro MA, Rojas M, Dominguez S, Vergara A. Cytokeratin, vimentin and E-cadherin immunodetection in the embryonic palate in two strains of mice with different susceptibility to glucocorticoid-induced clefting. J Craniofac Genet Dev Biol 2000;20:137–143.
Bartlett JD, Dobeck JM, Tye CE, et al. Targeted p120-catenin ablation disrupts dental enamel development. PloS One 2010;5:e12703.
Li CY, Cha W, Luder HU, et al. E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev Biol 2012;366:357–366.
Shimizu Y, Thumkeo D, Keel J, et al. ROCK-I regulates closure of the eyelids and ventral body wall by inducing assembly of actomyosin bundles. J Cell Biol 2005;168:941–953.
Smith AL, Dohn MR, Brown MV, Reynolds AB. Association of Rho-associated protein kinase 1 with E-cadherin complexes is mediated by p120-catenin. Mol Biol Cell 2012;23:99–110.
Hansford S, Kaurah P, Li-Chang H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol 2015;1:23–32.
van der Post RS, Vogelaar IP, Carneiro F, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet 2015;52:361–374.
Benusiglio PR, Caron O, Consolino E, et al. Cleft lip, cleft palate, hereditary diffuse gastric cancer and germline mutations in CDH1. Int J Cancer 2013;132:2470.
Kluijt I, Siemerink EJ, Ausems MG, et al.; Dutch Working Group on Hereditary Gastric Cancer. CDH1-related hereditary diffuse gastric cancer syndrome: clinical variations and implications for counseling. Int J Cancer 2012;131:367–376.
Brito LA, Yamamoto GL, Melo S, et al. Rare variants in the epithelial cadherin gene underlying the genetic etiology of nonsyndromic cleft lip with or without cleft palate. Hum Mutat 2015;36:1029–1033.
Vogelaar IP, Figueiredo J, van Rooij IA, et al. Identification of germline mutations in the cancer predisposing gene CDH1 in patients with orofacial clefts. Hum Mol Genet 2013;22:919–926.
Cadwell CM, Su W, Kowalczyk AP. Cadherin tales: Regulation of cadherin function by endocytic membrane trafficking. Traffic 2016;17:1262–1271.
Ciesiolka M, Delvaeye M, Van Imschoot G, et al. p120 catenin is required for morphogenetic movements involved in the formation of the eyes and the craniofacial skeleton in Xenopus. J Cell Sci 2004;117(Pt 18):4325–4339.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760.
DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498.
Li H, Handsaker B, Wysoker A, et al.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–2079.
Geoffroy V, Pizot C, Redin C, et al. VaRank: a simple and powerful tool for ranking genetic variants. PeerJ 2015;3:e796.
Miranda KC, Khromykh T, Christy P, et al. A dileucine motif targets E-cadherin to the basolateral cell surface in Madin-Darby canine kidney and LLC-PK1 epithelial cells. J Biol Chem 2001;276:22565–22572.
Tepass U, Truong K, Godt D, Ikura M, Peifer M. Cadherins in embryonic and neural morphogenesis. Nat Rev Mol Cell Biol 2000;1:91–100.
Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol 2012;22:299–310.
Harrison OJ, Jin X, Hong S, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 2011;19:244–256.
Reynolds AB, Roczniak-Ferguson A. Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 2004;23:7947–7956.
Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998;23:198–199.
Nishi E, Masuda K, Arakawa M, et al. Exome sequencing-based identification of mutations in non-syndromic genes among individuals with apparently syndromic features. Am J Med Genet A 2016;170:2889–2894.
Figueiredo J, Söderberg O, Simões-Correia J, Grannas K, Suriano G, Seruca R. The importance of E-cadherin binding partners to evaluate the pathogenicity of E-cadherin missense mutations associated to HDGC. Eur J Hum Genet 2013;21:301–309.
Benusiglio PR, Colas C, Rouleau E, et al. Hereditary diffuse gastric cancer syndrome: improved performances of the 2015 testing criteria for the identification of probands with a CDH1 germline mutation. J Med Genet 2015;52:563–565.
Petridis C, Shinomiya I, Kohut K, et al. Germline CDH1 mutations in bilateral lobular carcinoma in situ. Br J Cancer 2014;110:1053–1057.
Huynh JM, Laukaitis CM. Panel testing reveals nonsense and missense CDH1 mutations in families without hereditary diffuse gastric cancer. Mol Genet Genomic Med 2016;4:232–236.
Simões-Correia J, Figueiredo J, Oliveira C, et al. Endoplasmic reticulum quality control: a new mechanism of E-cadherin regulation and its implication in cancer. Hum Mol Genet 2008;17:3566–3576.
Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol 2003;163:525–534.
Tinkle CL, Lechler T, Pasolli HA, Fuchs E. Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc Natl Acad Sci USA 2004;101:552–557.
Acknowledgements
This work was supported by funding from the French Foundation for Rare Diseases (Fondation Maladies Rares). We thank Jerome Sige and Emilie Aït-Yahya-Graison, Plateforme de Bioinformatique, CHRU Lille, France, for their helpful support with exome data analysis. We also thank Myriem Tardivel, Plateforme d’Interaction Moléculaire, IMPRT-IFR114, Faculté de Médecine de Lille, France, for her helpful support with fluorescence microscopy imaging.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghoumid, J., Stichelbout, M., Jourdain, AS. et al. Blepharocheilodontic syndrome is a CDH1 pathway–related disorder due to mutations in CDH1 and CTNND1. Genet Med 19, 1013–1021 (2017). https://doi.org/10.1038/gim.2017.11
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.11
Keywords
This article is cited by
-
The European Journal of Human Genetics is turning 30: a selection of major cancer genetics papers published by the Journal
European Journal of Human Genetics (2022)
-
Pathogenic variants in CDH11 impair cell adhesion and cause Teebi hypertelorism syndrome
Human Genetics (2021)
-
Cleft lip/palate and hereditary diffuse gastric cancer: report of a family harboring a CDH1 c.687 + 1G > A germline mutation and review of the literature
Familial Cancer (2019)
-
Variants in members of the cadherin–catenin complex, CDH1 and CTNND1, cause blepharocheilodontic syndrome
European Journal of Human Genetics (2018)
-
CDH1 germline mutations: different syndromes, same management?
Genetics in Medicine (2017)