Abstract
Purpose:
Lynch syndrome (LS), a heritable disorder with an increased risk of primarily colorectal cancer (CRC) and endometrial cancer (EC), can be caused by mutations in the PMS2 gene. We wished to establish whether genotype and/or parent-of-origin effects (POE) explain (part of) the reported variability in severity of the phenotype.
Methods:
European PMS2 mutation carriers (n = 381) were grouped and compared based on RNA expression and whether the mutation was inherited paternally or maternally.
Results:
Mutation carriers with loss of RNA expression (group 1) had a significantly lower age at CRC diagnosis (51.1 years vs. 60.0 years, P = 0.035) and a lower age at EC diagnosis (55.8 years vs. 61.0 years, P = 0.2, nonsignificant) compared with group 2 (retention of RNA expression). Furthermore, group 1 showed slightly higher, but nonsignificant, hazard ratios (HRs) for both CRC (HR: 1.31, P = 0.38) and EC (HR: 1.22, P = 0.72). No evidence for a significant parent-of-origin effect was found for either CRC or EC.
Conclusions:
PMS2 mutation carriers with retention of RNA expression developed CRC 9 years later than those with loss of RNA expression. If confirmed, this finding would justify a delay in surveillance for these cases. Cancer risk was not influenced by a parent-of-origin effect.
Genet Med 18 4, 405–409.
Similar content being viewed by others
Main
A germ-line mutation in one of the mismatch repair (MMR) genes causes Lynch syndrome (LS), an autosomal dominant disorder characterized by the clustering of colorectal cancer (CRC) and endometrial cancer (EC) within affected families. Higher risks have been reported for other cancers such as ovarian and urothelial cell cancer. However, thus far only one study has confirmed these risks in PMS2 mutation carriers.1
The MMR proteins normally act together to repair mismatches that occur during cell replication. MSH2 and MSH6 form a heterodimer that recognizes base–base mismatches and insertion/deletion mispairs, whereas MLH1 and PMS2 form a heterodimer that supports initiated repair.2 A mutation can result in complete loss of protein or a protein with impaired function. Cancer risks associated with PMS2 are lower than those reported for MLH1 and MSH2.1,3
Phenotypes resulting from germ-line MMR gene mutations vary both among and within families.4 Interfamilial variance might be partly attributable to known genotype–phenotype correlations of MMR genes,5,6,7 whereas intrafamilial variance could be due to the influence of parental transmission on penetrance of the disease, a so-called parent-of-origin effect (POE). Although a POE has previously been reported in LS, studies have shown conflicting results.8,9
The aims of our study were to investigate genotype–phenotype relationships in PMS2 mutation carriers and to explore a possible parent-of-origin effect in PMS2. Significant results would have implications for the surveillance and management of patients and their families.
Materials and Methods
Patients
The study cohort included 381 presymptomatic and symptomatic mutation carriers (from 130 families) with a heterozygous PMS2 mutation and consisted of 120 apparently unrelated index patients and 261 relatives. Index cases with missing clinical data (n = 1) or constitutional mismatch repair deficiency (CMMR-D) (n = 9)10 were excluded from analysis (due to a younger age of onset and a different tumor spectrum compared with heterozygous mutation carriers, CMMR-D is considered a separate syndrome). When available, clinical data from participating clinical genetics departments (the Netherlands, Spain, Norway, Denmark, Sweden, and Germany) were used to confirm the diagnosis. DNA analysis of patients and family members was conducted between 2007 and 2013, and in the majority of cases the analysis was indicated due to compliance with the Bethesda criteria11 or MSI-testing-indicated-by-a-pathologist (MIPA) criteria.12 When applicable, informed consent was obtained according to local approved protocols (LUMC Ethics Review Board, P01.019). Information on inheritance was available for 183 PMS2 mutation carriers.
PMS2 mutations
The PMS2 mutations included in this study were detected using a range of mutation detection strategies as applied by the diagnostic laboratories connected to the aforementioned clinical genetic departments. All laboratories aimed at avoiding interference by pseudogenes by applying different methods (Supplementary Table S1 online). Data on RNA splicing and transcript expression were available for approximately half of all mutations13,14,15,16,17 The mutations were therefore classified into three genotype groups:
-
1. Mutations with an observed reduction in mRNA expression or entirely absent expression in RT-PCR screenings assays
-
2. Mutations without an effect on mRNA expression
-
3. Mutations for which no or inconclusive data on RNA analysis were available
PMS2 mutations were described according to Human Genetic Variation Society–approved guidelines (http://www.hgvs.org/mutnomen/) ( Table 1 and Supplementary Table S1 online). Group 1 included a missense, c.903G>T (p.Tyr268), and a silent mutation, c.825A>G (p.Ile269Alafs*31), with a known effect on splicing, large genomic out-of-frame deletions, and deletions that involve the start and/or the end of the gene. Group 2 consisted of two missense mutations, 137G>T (p.Ser46Ile) and c.2113G>A (p.Glu705Lys), that were shown to be mismatch-repair–deficient in a cell-free functional test.18 Although no functional assay was available for a third missense mutation, c.2444C>T (p.Ser815Leu), analysis with three in silico prediction programs classified this variant as deleterious (SIFT; score 0.00), probably damaging (Polyphen-2; score 1.00), and likely to interfere with function (aGVGD; class C65).
Group 3 consisted of all mutations for which protein expression from the mutated allele was uncertain, including large genomic in-frame deletions, splice variants causing in-frame exon skipping (e.g., exon 10 deletion or skip), splice variants inducing multiple aberrant transcripts (of which some are in-frame), and nonsense or frame-shift mutations that escape NMD due to their location in the gene.
Statistics
The chi-square test and the Cox regression analyses were performed separately for CRC and EC using IBM SPSS Statistics 20. A P value <0.05 was considered to be statistically significant. Because the majority of group 3 mutations probably result in loss of RNA expression, mutation carriers in groups 1 and 3 were combined and then compared with group 2 in a sensitivity analysis.
For the CRC risk analyses, using the Cox regression method, mutation carriers were considered to be informative from birth until complete or partial colectomy, start of surveillance and/or first polyp detection, last contact, or death. In the case of the EC risk analyses, mutation carriers were considered to be informative until hysterectomy, last contact, or death. The development of CRC or EC was considered the end point. Mutation carriers could reasonably be considered informative from birth because a very young age of cancer development would prompt genetic testing, resulting in no young index case being missed. These analyses resulted in hazard ratios comparing the two groups.
Generalized estimating equations were used to rule out the possibility that Cox regression results were influenced by the coincidental clustering of family characteristics other than the mutation itself.
Results
In 381 mutation carriers from 130 families, a total of 53 different PMS2 germ-line mutations were found, with 248 mutation carriers carrying a mutation that results in loss of RNA expression. The most common mutations were c.736_741delCCCCTinsTCTCTCTGAAG, present in 61 mutation carriers, and c.1882C>T, present in 47 mutation carriers. Together, these two mutations accounted for 28.3% of all mutation carriers (Supplementary Table S1 online).
Genotype groups
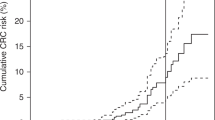
Of the 282 mutation carriers in RNA groups 1 and 2, 96 developed CRC. A significant difference (P = 0.035) was noted in mean ages of CRC development (51.1 vs. 60.0 years) ( Table 1 ). Comparing groups 1 and 2 produced no evidence for a significantly elevated risk of CRC development (HR: 1.31, P = 0.38). Of the 155 women included in the analyses, 27 developed EC, with a slightly lower, but nonsignificant, mean age of EC development in group 1 compared with group 2 (55.8 years vs. 61.0, P = 0.2).
Evidence that males have a higher risk of developing an MMR-related CRC prompted us to include gender as a co-variate.9,11 As expected, males had a higher risk (HR: 1.72) of developing CRC than females (P = 0.012). No statistically significant differences in EC development were found for the 140 women in the various genotype groups (HR: 1.22, P = 0.72).
Sensitivity analyses
For this analysis, mutation carriers in group 1 were combined with mutation carriers in group 3. This new group of mutation carriers was then compared with mutation carriers in group 2. The HR for CRC development (HR: 1.30, P = 0.39) was similar to the HR for just groups 1 and 2. The HR for EC was slightly lower (HR: 1.07, P = 0.91). Additional analysis using generalized estimating equations showed no significant differences for the different genotype groups after clustering the data.
Parent of origin
The parent of origin was known for 183 of the 381 mutation carriers. Of these, 39 developed CRC and 9 females developed EC. Inheritance of a mutation via either the paternal or the maternal line did not significantly influence the mean age of CRC (46.9 vs. 45.6 years, P = 0.68) or EC onset (49.2 vs. 55.5 years, P = 0.23). Cox regression analysis, in both the group as a whole and following separate analysis of males and females, also produced no evidence for a POE. The HR for CRC associated with paternal inheritance of mutations was comparable to that for maternal inheritance (0.80, P = 0.51). For EC, the hazard ratio was 1.73 (P = 0.46; Table 2 ).
Discussion
Mutation carriers with a PMS2 mutation that results in the loss of RNA expression develop CRC 9 years earlier than carriers of mutations that do not effect RNA expression. An explanation for this finding could be that, in the latter group, protein with some residual function is still produced. Although some (functional) protein expression (~25%)19 and a (limited) repair function was found for the c.2113G>A mutation, functional studies demonstrated severely impaired repair efficiency,18,19 perhaps suggesting that a severely impaired protein is still superior to no protein at all.
Further support for the notion that mutations leading to retention of mRNA expression result in a milder phenotype comes from the underreporting of this type of mutation. Both in the present study and that of Senter et al.,3 the majority of PMS2 mutations found in patients result in loss of RNA expression. On a population basis, there is no obvious reason that fewer group 2 than group 1 mutations would occur, suggesting that individuals with group 2 mutations may have less severe phenotypes and/or no family history and are therefore less likely to be referred to a clinical geneticist. This idea was previously suggested by Beck et al.,20 who found a relative overabundance of missense MLH1 and MSH2 mutations in 10 families that failed to meet the Amsterdam I criteria compared with families that meet the criteria.
There are a number of shortcomings of our study. For one, the inclusion of affected family members of index patients might have resulted in bias due to the phenotypes of relatives being more similar than phenotypes of unrelated index cases. We attempted to overcome this problem using generalized estimating equations analysis. We also repeated the analyses for index patients alone, which resulted in similar HR and P values ( Table 2 ). Another shortcoming was the relatively small number of patients in group 2, reducing statistical power. This may explain nonsignificant results and indicates the need for analyses in larger patient groups. Also in this group, one mutation, the c.2444C>T, found in one family, is classified as VUS/Class 3 in the InSight database (link: http://insight-group.org/variants/database/), and larger cohorts are still needed to prove its pathogenicity. When excluding the family (four relatives) from the analyses, similar HR and P values were found ( Table 2 ) and a significant difference in age of CRC development remained (51.1 years in group 1 vs. 61.5 years in group 2; P = 0.17). Finally, two mutations are overrepresented in our cohort (namely the c.736_741delinsTGTGTGTGAAG and the c.1882C>T mutation) and might dominate the results. However, when comparing patients with these mutations and patients with other mutations, no significant differences in mean age of CRC or CRC risk (HR) were found.
We were unable to confirm earlier reports that found a significant POE.8,9 However, a trend was observed toward a lower HR for CRC in females with a paternally inherited mutation. This is broadly in line with the results of van Vliet et al.8 for the males in their research population, although their results showed a much higher, and significant, HR of 3.2 (P = 0.03) for males when comparing maternally inherited mutations with paternally inherited mutations.
A possible explanation for the differences in POE findings could be the fact that van Vliet et al.8 used another statistical approach—a modified segregation analyses. We did not use this broad approach because, to the best of our knowledge, no bias or confounders were present in our cohort that would make a modified segregation analysis necessary. The possible exception would be a POE-dependent selection bias; for example, if mutation carriers with a maternally inherited mutation were more severely affected, more carriers of a maternally inherited mutation with CRC would be expected in our database. Using a chi-square test, we therefore analyzed whether there was a bias in maternal inheritance for mutation carriers with CRC compared with those without CRC. This was not the case (P = 0.12).8,9
With the ever-wider adoption of whole genome DNA analysis, more families with PMS2 mutations will be identified in the near future, including some with no apparent history suggestive of LS. Because many of these families may have milder phenotypes, studies such as ours provide useful advice on surveillance programs for these mutation carriers. Should our results be confirmed in larger studies, the significant age differences in CRC development reported here provide some justification for starting surveillance at a later age for mutation carriers who show retention of PMS2 RNA expression.
Disclosure
G.C.M. owns stock in, has leadership and consulting/advisory roles with, and has received honoraria from VCN Biosciences. N.H. and A.M. hold consulting/advisory roles with AstraZeneca. The other authors declare no conflict of interest.
References
ten Broeke SW, Brohet RM, Tops CM, et al. Lynch syndrome caused by germline PMS2 mutations: delineating the cancer risk. J Clin Oncol 2015;33:319–325.
Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem 2006;281:30305–30309.
Senter L, Clendenning M, Sotamaa K, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology Aug 2008;135(2):419–428.
Kohlmann W, Gruber SB .Lynch Syndrome. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, eds. GeneReviews. Seattle,WA, 1993.
Peltomäki P, Gao X, Mecklin JP. Genotype and phenotype in hereditary nonpolyposis colon cancer: a study of families with different vs. shared predisposing mutations. Fam Cancer 2001;1:9–15.
Pérez-Cabornero L, Infante M, Velasco E, Lastra E, Miner C, Durán M. Genotype-phenotype correlation in MMR mutation-positive families with Lynch syndrome. Int J Colorectal Dis 2013;28:1195–1201.
Geary J, Sasieni P, Houlston R, et al. Gene-related cancer spectrum in families with hereditary non-polyposis colorectal cancer (HNPCC). Fam Cancer 2008;7:163–172.
van Vliet CM, Dowty JG, van Vliet JL, et al. Dependence of colorectal cancer risk on the parent-of-origin of mutations in DNA mismatch repair genes. Hum Mutat 2011;32:207–212.
Green J, O’Driscoll M, Barnes A, et al. Impact of gender and parent of origin on the phenotypic expression of hereditary nonpolyposis colorectal cancer in a large Newfoundland kindred with a common MSH2 mutation. Dis Colon Rectum 2002;45:1223–1232.
Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg? Hum Genet 2008;124:105–122.
Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004;96:261–268.
Overbeek LI, Hermens RP, van Krieken JH, et al.; MIPA study group. Electronic reminders for pathologists promote recognition of patients at risk for Lynch syndrome: cluster-randomised controlled trial. Virchows Arch 2010;456:653–659.
Borràs E, Pineda M, Cadiñanos J, et al. Refining the role of PMS2 in Lynch syndrome: germline mutational analysis improved by comprehensive assessment of variants. J Med Genet 2013;50:552–563.
Johannesma PC, van der Klift HM, van Grieken NC, et al. Childhood brain tumours due to germline bi-allelic mismatch repair gene mutations. Clin Genet 2011;80:243–255.
Sjursen W, Bjørnevoll I, Engebretsen LF, Fjelland K, Halvorsen T, Myrvold HE. A homozygote splice site PMS2 mutation as cause of Turcot syndrome gives rise to two different abnormal transcripts. Fam Cancer 2009;8:179–186.
van der Klift HM, Tops CM, Bik EC, et al. Quantification of sequence exchange events between PMS2 and PMS2CL provides a basis for improved mutation scanning of Lynch syndrome patients. Hum Mutat 2010;31:578–587.
van der Klift HM, Tops CM, Hes FJ, Devilee P, Wijnen JT. Insertion of an SVA element, a nonautonomous retrotransposon, in PMS2 intron 7 as a novel cause of Lynch syndrome. Hum Mutat 2012;33:1051–1055.
Drost M, Koppejan H, de Wind N. Inactivation of DNA mismatch repair by variants of uncertain significance in the PMS2 gene. Hum Mutat 2013;34:1477–1480.
Deschênes SM, Tomer G, Nguyen M, et al. The E705K mutation in hPMS2 exerts recessive, not dominant, effects on mismatch repair. Cancer Lett 2007;249:148–156.
Beck NE, Tomlinson IP, Homfray T, Hodgson SV, Harocopos CJ, Bodmer WF. Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal cancer even if the Amsterdam criteria are not fulfilled. Br J Surg 1997;84:233–237.
Acknowledgements
The authors thank Medactie.com for help with the writing and editing of this paper. This research was funded by a grant from the Dutch Cancer Society (KWF UL 2012–5155).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Table S1
(DOC 103 kb)
Rights and permissions
About this article
Cite this article
Suerink, M., van der Klift, H., ten Broeke, S. et al. The effect of genotypes and parent of origin on cancer risk and age of cancer development in PMS2 mutation carriers. Genet Med 18, 405–409 (2016). https://doi.org/10.1038/gim.2015.83
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2015.83
Keywords
This article is cited by
-
SNP association study in PMS2-associated Lynch syndrome
Familial Cancer (2018)