Abstract
A large part of Y chromosome lineages in East European and East Asian human populations belong to haplogroup (hg) NO, which is composed of two sister clades N-M231 and O-M175. The O-clade is relatively old (around 30 thousand years (ky)) and encompasses the vast majority of east and Southeast Asian male lineages, as well as significant proportion of those in Oceanian males. On the other hand, our detailed analysis of hg N suggests that its high frequency in east Europe is due to its more recent expansion westward on a counter-clock northern route from inner Asia/southern Siberia, approximately 12–14 ky ago. The widespread presence of hg N in Siberia, together with its absence in Native Americans, implies its spread happened after the founder event for the Americas. The most frequent subclade N3, arose probably in the region of present day China, and subsequently experienced serial bottlenecks in Siberia and secondary expansions in eastern Europe. Another branch, N2, forms two distinctive subclusters of STR haplotypes, Asian (N2-A) and European (N2-E), the latter now mostly distributed in Finno-Ugric and related populations. These phylogeographic patterns provide evidence consistent with male-mediated counter-clockwise late Pleistocene–Holocene migratory trajectories toward Northwestern Europe from an ancestral East Asian source of Paleolithic heritage.
Similar content being viewed by others
Introduction
The global variation of the largely non-recombining male-specific Y chromosome has become one of the major sources in reconstructing ancient human migrations. The resolution of the Y chromosome phylogeny has increasingly been improved by the discovery of new binary polymorphisms, mostly single nucleotide polymorphisms (SNPs), that may, through their distinct geographic patterns, bear evidence of historic relationships between living populations.1, 2, 3, 4, 5
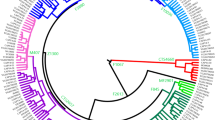
One of the most widespread and frequent branches of the Y phylogeny in Eurasia is haplogroup (hg) NO defined by SNP-marker M2145 (corrected phylogeny in Cinnioglu et al6) (see Figure 1). It entails a low number of NO* lineages that lack distinguishing derived SNP markers (Figure 2a) and two frequent sister clades, N and O, defined by markers M231 and M175, respectively (Figure 2b,c). Although the phylogeography of clade O has drawn considerable scrutiny,7, 8, 9, 10 knowledge about hg N is relatively impoverished with regards to its origin, phylogeographic patterning and demographic significance.
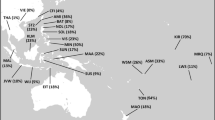
Geographical distribution of NO clade. (a–g) Spatial frequency distributions of the NO clade: NO*, N (overall distribution of hg N), O (overall distribution of hg O), N*, N1, N2, N3. Maps are based on data from Supplementary Table 1. We label various panels following the YCC ‘by mutation’ format by adding the relevant mutation suffix.
Materials and methods
Samples and DNA typing
A total of 5389 samples from 58 populations in different geographical regions were genotyped or updated to present phylogenetic resolution (M9-derived samples with ancestral allele of 92R7 marker were typed for M214, M231, M128, P43, Tat, M175) in this study and analyzed together with data about 8019 individuals from 90 populations from the literature (data presented in Supplementary Table 1). DNA samples were obtained from unrelated male volunteers after getting the informed consent from ethical committees of institutions involved.
Mutation labeling follows the YCC nomenclature.1, 4 Phylogenetic relations of markers M128, P43 and TAT, characterizing three subclades (N1–N3 respectively) were known earlier,2, 11, 12 but only recently marker M2316 (characterizing the whole N clade) was introduced in the tree of Y chromosome diversification (see Figure 1). Marker M231 is phylogenetically equivalent to the more cumbersome LLY22g polymorphism,13 initially used to define haplogroup N.
Markers M128 and M214 2, 5 were assayed by sequencing the polymorphic sites, and markers M175,2 P4311 and Tat12 were assayed using restriction-fragment length polymorphism (RFLP) method using restriction enzymes MboII, NlaIII and TaiI, respectively. The allelic state of the M231, SNP first described in Cinnioglu et al6 and originally assayed by denaturing high performance liquid chromatography method, can be readily assayed by RFLP analysis (TaqI enzyme cuts ancestral allele G, producing 223 and 108 bp products and does not cut derived allele A with lenght of 331 bp).
STRs were studied using Y-filer Kit (Applied Biosystems, Foster City, CA, USA). PCR products were analyzed on ABI 3100Avant genetic analyzer (Applied Biosystems) in the mode of standard fragment analysis protocol. GeneScan 500LIZ size standard (Applied Biosystems) was added to each sample for size scaling, and GeneMapper 3.5 (Applied Biosystems) was employed for allele scoring. Alleles were designated by repeat numbers.
Data analysis
Using the program Network 4.1.1.2., median joining network was constructed of hg N–O haplotypes from data on 17 STRs (DYS19, DYS385a,b, DYS389I,II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYs635, Y GATA) and bi-allelic markers (M231, M128, P43, TAT) in 58 individuals (STR data presented in Supplementary Table 2). Phylogenetic relationships between the haplotypes were determined by the median joining method after having processed the data with the reduced median method described in Bandelt et al14 using program Network 4.1.1.2, http://www.fluxus-engineering.com.
The age calculations are based on STR variation and calculated according to published methods.15, 16
Spatial frequency maps of NO clade and subclades were obtained applying the frequencies from Supplementary Table 1 (dots indicate the populations) in Surfer software (version 7, Golden Software, Inc.).
Results and discussion
Different distribution-pattern of N2 and N3 versus NO*, O and N*
Haplogroup N has both a unique and widespread distribution spanning northern Eurasia, from the Far East to Eastern Europe, showing higher frequencies at high latitudes.11, 12, 17 Here, we assess the history of this haplogroup via a detailed phylogeographic approach using samples from different regions of Europe, East/Southeast Asia and Oceania, ascertaining SNP markers defining haplogroup N, its subclades and sister-clade O. The analysis reveal that despite its ancient split from hg O, hg N subclades display more recent demographic temporality and a net counter-clockwise migratory trajectory distinctive from its hg O counterparts.
Although having variable frequency scales, the spatial distributions for ancestral paragroup NO-M214*, paragroup N-M231* and the prevalent hg O-M175 (Figure 2a, c, d) are generally congruent and highlight Southeast Asia as the most parsimonious source region of these clades. The spread pattern of paragroup NO* approximates the same regions of Southeast Asia as paragroup N*, although being present at an even lower frequency compared with N*18, 19 (data from Kayser et al19 updated in present study). More notable, however, is the fact that the spatial dynamics of the whole N and O haplogroups greatly differ from each other. The split between N* and O is dated to 34.6±4.7 thousand years (ky). The age of STR variation of hg O in Southeast Asia probably exceeds 26 ky,10 and its numerous subclades currently predominate in southern and southeastern Asia extending into northern China, Manchuria and some Siberian populations,7, 9, 11, 20, 21 as well as westward to the eastern sector of the Indian subcontinent10 and eastward to Oceania.18, 19
Distribution and spread of haplogroup N subclades
The N-haplogroups reflect a more recent demographic history. Ancestral paragroup N* is widely distributed, although with low frequencies, from Fiji, Borneo, Cambodia, southern China and Japan up to southern Siberia (Supplementary Table 1), while apparently absent in the Indian Peninsula.10 Its age of accumulated STR variation estimated using the method from Zhivotovsky et al15 points to late Pleistocene–early Holocene (11.9±2.5 to 12.6±3.1 ky), depending on the number of Y chromosomes and STR loci included in analysis (see Table 1). However, it should be noticed that the frequency of N* is extremely low.
In this regard, the age of accumulated STR variation in hg N, estimated on all combined data from N1, N2 and N3 at 15 loci (Supplementary Table 2), yields an estimate of 19.4±4.8 ky. However, as will be argued below, the European subcluster of N2 and the Yakutian N3 might have descended from single founders with multiple jumps at several loci, thus causing a possible shift in statistical estimates that assume a step-wise mutation model. When those chromosomes are excluded, the age of hg N STR diversity is somewhat younger, 14.2±4.0 ky.
Time calculations based on evolutionary- and pedigree-based methods give significantly different date estimates (Table 1). Both estimates are included because a consensus has not yet been reached among all the geneticists. Recent simulations demonstrate that pedigree rates do not consider the evolutionary consequences of population dynamics, such as the rapid extinction of newly arisen microsatellite alleles (Zhivotovsky, Underhill and Feldman).23 Thus, time estimates based on pedigree studies are younger and inconsistent with the archaeological record. Additional factors relevant to the issue include (i) ascertainment bias (studies reporting no mutations in a pedigree are less likely to be published); (ii) rate variation between loci whereby pedigree rate yields the average rate of the fastest evolving loci; (iii) saturation (the evolutionary rate calibration misses back-and-forth mutations).
The median joining network (Figure 3), based on 17 STR loci (Supplementary Table 2), and SNPs shows the extent of variation within hg N subhaplogroups. Despite the current presence of N3 and N2 in various Siberian populations, including Chukchi and Yupik from Chukotka Peninsula in Beringia, these haplogroups are absent among Native Americans.24, 25, 26, 27, 28 This finding suggests that hg N chromosomes were likely not among the dominant and omnipresent types in Palaeolithic Siberians at the time of their likely colonization of the Americas some 12–17 ky,29 although the possibility that the N lineages became extinct during the colonization due to founder effect or drift cannot be excluded.
Median network of N–O haplogroups. Median joining network of hg N–O haplotypes was constructed based on data of 17 STRs and bi-allelic markers in 58 individuals by using the program Network 4.1.1.2. Each circle represents a haplotype, defined by a combination of STR markers. Circle size is shown proportional to haplotype frequency, according to data presented in Supplementary Table 2. Haplotypes are labeled as follows: Al–Altaian, Ba–Bashkir, Ch–Chinese, Ci–Chukci, Cu–Chuvash, Eo–Eskimo, Ee–Estonian, Ev–Evenk, Fj–Fiji, Ka–Karelian, Kh–Khakash, Ko–Komi, Ma–Mari, Ru–Russian, Sl–Slovak, Ta–Tatar, Tu–Tuva, Ud–Udmurt, Uk–Ukrainian, Vp–Vepsa, Ya–Yakut. Colors indicate the subdivisions inside haplogroups: light green for European N3-haplotypes, dark green for Asian N3-haplotypes, light blue for European N2-haplotypes, blue for Asian N2-haplotypes, brown for N1-haplotypes, pink for N* haplotypes, purple for O-haplotype.
Haplogroup N3 is the most common subclade of hg N (Figure 2g, Supplementary Table 1), being almost universally the most frequent Y chromosome type among populations inhabiting north Eurasia,11, 12, 17, 26, 30, 31, 32, 33, 34, 35 while occurring at only marginal frequencies in China, Korea, Borneo and Japan.18, 19 Being prevalent throughout northern Asia, its distribution in Europe is restricted to the northern and eastern populations, showing sharp east–west decline across Scandinavia and between Lithuania and Poland (Supplementary Table 1 and data in17). The phylogeography of the NO* and N* lineages (Figure 2a, d) and the presence of N* chromosomes in southern East Asia (South China and Cambodia, see Supplementary Table 1) suggests that this region could be the source of the initial spread of hg N. In this scenario, the Altay/Sayan/southern Siberia region might have been a place of transition of hg N westward as all major subclades of hg N are still to be found there.
Although the frequency of hg N3 is low in northern China and restricted to a few small populations, its STR variance is higher (0.26, averaged across eight loci: DYS19; DYS389Iⅈ DYS391; DYS392; DYS393 and DYS439, data from Sengupta et al10) than in Altai and in Volga-Ural region (0.16 and 0.17, respectively), thus again pointing to northern China rather than southern Siberia as a possible place of expansion of hg N3. The age of accumulated N3-STR variation in North China is 11.8±6.8 ky, falling, thus, at the boundary of Pleistocene and Holocene, although it should be treated with caution because of a very large standard error, caused by limited sample size of the N3 chromosomes.
According to our scenario, on the way through Siberia to eastern Europe, the N3-carriers might have been subjected to founder effects or strong genetic bottlenecks. Northeastern Europe can be considered as a place of secondary expansion of N3. Indeed, hg N3 occurs at high frequencies in the Volga-Ural Ugric groups and related Finns, Saami and Estonians. One may notice that while STR variation is relatively low in the Volga-Ural group, some north-European populations have high STR variance (eg, 0.32 in Finns: data from,36 without DYS385ab). The high STR variation among the latter, however, might not be a result of a long-term in situ differentiation of the founder lineage, but, rather a consequence of an admixture of separate N3 founder types.
Populations of eastern Europe on the most distant western border of N3 spread area that have considerable frequencies of hg N3 from single sources are expected to have lower STR variation. As an example, STR variance in Baltic-Lithuanians and Latvians is 0.12 and 0.09, respectively (data on five loci).37 Some European populations have low frequency of hg N3 combined with high STR variation as in non-Saami Norwegians (0.27: data without DYS439) that may indicate recent gene flow from the neighboring Finno-Ugric populations.36, 38 Similar situation for Swedes was described recently in study about Swedish Y-chromosomal pool.39
Phylogenetic analysis of STR variation (Figure 3) shows two overlapping subclusters of N3, one of them encompassing predominantly Volga-Ural region, Finnic- as well as Turkic-speaking populations together with Altaian, and the other one both Baltic-Finnic (Estonians, Karelians and Vepsa) and east Slavs (Russians, Ukrainians), as well as West-Slavonic Slovak N3 chromosomes. The Yakut Y chromosomes form their specific branch; they are almost identical to each other, consistent with earlier studies.12, 40
The haplogroup N2 distribution (Figure 2e) exhibits an irregular frequency pattern in Siberian populations, extending in the western direction to eastern Europe as far as Vepsas and Karelians at the Baltic Sea. The highest frequencies of N2 are observed among north-west Siberian populations: 92% in the Nganassan, 78% in the Enets and 74% in the Tundra Nenets.11 In Europe, the N2 types have their highest frequency of 20% among Volga-Uralic populations.17 The extreme western border of the spread of N2 is Finland, where this haplogroup occurs only at marginal frequency – 0.4%.36 Yet interestingly, N2 is quite frequent among Vepsas (17.9%), a small Finnic population living in immediate proximity to Finns, Karelians and Estonians.
The network of N2 haplotypes shows a well-resolved bipartite STR distribution with separate European and Siberian subclusters, denoted here by N2-A and N2-E (Figure 3). It can be speculated that the nearest Asian putative root subcluster, N2-A, originated first, later giving rise to the derived European subcluster, N2-E. Although the N2-A has median repeat scores more similar to those for N3, the European subcluster N2-E differs sharply from N2-A in its STR composition at several loci (Table 2), thus suggesting that the European N2-chromosomes descended from a single founding haplotype. One can even speculate on the probable existence of binary polymorphisms yet to be discovered which would be unique to the N2-E cluster.
N2-A and N2-E clusters are relatively young – the ages of accumulated STR variation in N2-A and N2-E are 6.2±2.0 and 6.8±2.9 ky, respectively; the lower value for presumably older hg N2-A can be explained by stronger bottlenecks in Siberian populations and by small sample sizes. Indeed, the indigenous Siberian populations are very small in size compared with most of east European populations; even the most numerous of the former, Yakuts and Buryats, reach only a few hundreds of thousands – compared with many millions of east Europeans.41, 42
Among our samples, N2-E is mainly restricted to the Volga-Ural region, which might be a possible source region for the northward and eastward (Khants and Mansis; data from Stepanov et al43) gene flow of N2. In contrast to NW-Siberian N2-A STR profile, the more western lineages are of the N2-E type. Interestingly, 14 N2-individuals from Turkey, data from Cinnioglu et al6 (updated in this study), belong to the Asian subcluster N2-A, suggesting that the clade N2 might have geographically expanded from Siberia westward by at least two different flows: one northwest through the Volga-Ural region, giving rise to N2-E, probably mainly via the Finno-Ugric group, and the other, N2-A, southwest together with Turkic languages. Therefore, the distinctive difference of N2-E from N2-A in their STR composition, as well as data on similarity of STR profiles at N3 in the Yakut, indicates the consequence of multiple postglacial founder events, especially in the re-peopling of sparsely inhabited territories, consistent with the view on Central Asia as the ‘land of bottlenecks’.44
The least frequent N subclade is N1 (Figure 2f), distributed with low frequencies in some Central Asian populations, Koreans, Northern Hans and Manchurian Evenks. Further large-scale studies on present phylogenetic resolution level (earlier literature often do not provide necessary data) are needed to say more of the spread and distribution pattern of this clade.
In summary, Y chromosome haplogroup N presents a case of gene flow to eastern Europe that has its likely ultimate source in east Asia. There are no equal mtDNA counterparts for the NRY hg N narrative – the mtDNA haplogroups characteristic to southeast Asian populations occur in east Baltics with their total frequency of less than 1%.17, 45, 46 Only some minor twigs of the Asian mtDNA tree, like Z1 and D5, having high diversity in Altai/Central Asia, occur at above 1% in some Nordic populations like Saami and Finns.17, 45, 47 However, numerous mtDNA haplogroups, such as B, C, D, F and G, do span from South China to Siberia and Central Asia, up to the Ural Mountains and, at already lower frequencies, to the Turkic and Finno-Ugric populations of the Volga basin.48, 49, 50, 51, 52, 53
Although the frequency scales of these haplogroups are significantly different across different loci, this independent evidence provided by maternal ancestry supports significant pre-historic migration of humans from southeast Asia, back to the West via the counter-clockwise northern route.
References
Jobling MA, Tyler-Smith C : The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 2003; 4: 598–6123.
Underhill PA, Shen P, Lin AA et al: Y chromosome sequence variation and the history of human populations. Nat Genet 2000; 26: 358–361.
Hammer MF, Karafet TM, Redd AJ et al: Hierarchical patterns of global human Y-chromosome diversity. Mol Biol Evol 2001; 18: 1189–1203.
Y Chromosome Consortium: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Underhill PA, Passarino G, Lin AA et al: The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 2001; 65: 43–62.
Cinnioglu C, King R, Kivisild T et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Su B, Jin L : Natives or immigrants: modern human origin in East Asia. Nat Rev 2000; 1: 126–133.
Wen B, Li H, Lu D et al: Genetic evidence supports demic diffusion of Han culture. Nature 2004; 431: 302–305.
Shi H, Dong YL, Wen B et al: Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet 2005; 77: 408–419.
Sengupta S, Zhivotovsky LA, King R et al: Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of central asian pastoralists. Am J Hum Gen 2006; 78: 202–221.
Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF : High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol 2002; 74: 761–789.
Zerjal T, Dashnyam B, Pandya A et al: Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 1997; 60: 1174–1183.
Hurles ME, Sykes BC, Jobling MA, Forster P : The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am J Hum Genet 2005; 76: 894–901.
Bandelt HJ, Macaulay V, Richards M : Median Networks: Speedy Construction and Greedy Reduction, One Simulation, and Two Case Studies from Human mtDNA. Mol Phylogenet Evol 2000; 16: 8–28.
Zhivotovsky LA, Underhill PA, Cinnioglu C et al: On the effective mutation rate at Y-chromosome STRs with application to human population divergence time. Am J Hum Genet 2004; 74: 50–61.
Dupuy BM, Stenersen M, Egeland T, Olaisen B : Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum Mutat 2004; 23: 117–124.
Tambets K, Rootsi S, Kivisild T et al: The western and eastern roots of the Saami – the story of genetic ‘outliers’ told by mtDNA and Y-chromosome. Am J Hum Genet 2004; 74: 661:682.
Hammer MF, Karafet TM, Park H et al: Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J Hum Genet 2006; 51: 47–58.
Kayser M, Brauer S, Weiss G et al: Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am J Hum Genet 2003; 72: 281–302.
Su B, Xiao J, Underhill P et al: Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last ice age. Am J Hum Genet 1999; 65: 1718–1724.
Tajima A, Pan IH, Fucharoen G et al: Three major lineages of Asian Y chromosomes: implications for the peopling of east and southeast Asia. Hum Genet 2002; 110: 80–88.
Rootsi S, Magri C, Kivisild T et al: Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe. Am J Hum Genet 2004; 75: 128–137.
Zhivotovsky LA, Underhill PA, Feldman MW : Difference between evolutionarily effective and germ-line mutation rate due to stochastically varying haplogroup size. Mol Biol Evol 2006; 23: 2268–2270.
Bergen AW, Wang CY, Tsai J et al: An Asian-Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet 1999; 63: 63–80.
Karafet TM, Zegura SL, Posukh O et al: Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 1999; 64: 817–831.
Lell JT, Sukernik RI, Starikovskaya YB et al: The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet 2002; 70: 192–206.
Seielstad M, Yuldasheva N, Singh N et al: A novel Y-chromosome variant puts an upper limit on the timing of first entry into the Americas. Am J Hum Genet 2003; 73: 700–705.
Bortolini MC, Salzano FM, Thomas MG et al: Y-chromosome evidence for differing ancient demographic histories in the Americas. Am J Hum Genet 2003; 73: 524–539.
Zegura SL, Karafet TM, Zhivotovsky LA, Hammer MF : High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol Biol Evol 2004; 21: 164–175.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: A Y chromosome perspective. Science 2000; 290: 1155–1159.
Zerjal T, Beckman L, Beckman G et al: Geographical, linguistic, and cultural influences on genetic diversity: Y-chromosomal distribution in Northern European populations. Mol Biol Evol 2001; 18: 1077–1087.
Rootsi S, Kivisild T, Tambets K et al: On the phylogeographic context of sex-specific genetic markers of Finno-Ugric populations; in Künnap A (ed): The roots of peoples and languages of Northern Eurasia II and III. Tartu: University of Tartu. Division of Uralic Languages/Societas Historiae Fenno-Ugricae, 2000, pp 148–164.
Rosser ZH, Zerjal T, Hurles ME et al: Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 2000; 67: 1526–1543.
Villems R, Rootsi S, Tambets K et al: Archaeogenetics of Finno-Ugric speaking populations; in Julku K (ed):: The Roots of Peoples and Languages of Northern Eurasia IV. Oulu: Societas Historiae Fenno-Ugricae, 2002, pp 271–284.
Laitinen V, Lahermo P, Sistonen P, Savontaus ML : Y-chromosomal diversity suggests that Baltic males share common Finno-Ugric-speaking forefathers. Hum Hered 2002; 53: 68–78.
Lappalainen T, Koivumki S, Salmela E et al: Regional differences among the Finns: A Y-chromosomal perspective. Gene 2006; 376: 207–215.
Kasperavičiūtè D, Kučinskas V, Stoneking M : Y chromosome and mitochondrial DNA variation in Lithuanians. Ann Hum Genet 2004; 68: 438–452.
Dupuy BM, Stenersen M, Lu TT, Olaisen B : Geographical heterogeneity of Y-chromosomal lineages in Norway. Forensic Sci Int 2005; Epub. December 6.
Karlsson AO, Wallerstrom T, Gotherstrom A, Holmlund G : Y-chromosome diversity in Sweden – A long-time perspective. Eur J Hum Genet 2006, May 24 (epub).
Pakendorf B, Morar B, Tarskaia LA et al: Y-chromosomal evidence for a strong reduction in male population size of Yakuts. Hum Genet 2002; 110: 198–200.
Kolga M, Tõnurist I, Vaba L, Viikberg J : The Red Book of the Peoples of the Russian Empire. Tallinn, NGO Red Book, 2001.
Russian Population Census. 2002, www.perepis2002.ru.
Stepanov VA, Kharkov VN, Puzyrev VP : Evolution and phylogeography of human Y-chromosomal lineages (in Russian). Vestnik VOGiS 2006; 10: 57–73.
Zerjal T, Wells R, Yuldasheva N, Ruzibakiev R, Tyler-Smith C : A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet 2002; 71: 466–482.
Richards M, Macaulay V, Hickey E et al: Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 2000; 67: 1251–1276.
Pliss L, Tambets K, Loogväli EL et al: Mitochondrial DNA Portrait of Latvians: Towards the Understanding of the Genetic Structure of Baltic-Speaking Populations. Ann Hum Genet 2005; 69: 1–20.
Finnila S, Lehtonen MS, Majamaa K : Phylogenetic network for European mtDNA. Am J Hum Genet 2001; 68: 1475–1484.
Torroni A, Sukernik RI, Schurr TG et al: mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet 1993; 53: 591–608.
Comas D, Plaza S, Wells RS, Yuldaseva N, Lao O, Calafell F, Bertranpetit J : Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages. Eur J Hum Genet 2004; 12: 495–504.
Bermisheva M, Tambets K, Villems R, Khusnutdinova E : Diversity of mitochondrial DNA haplotypes in ethnic populations of the Volga-Ural region of Russia. Mol Biol 2002; 36: 990–1001.
Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP : Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 2002; 70: 635–651.
Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R : The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 2002; 19: 1737–1751.
Kong QP, Bandelt HJ, Sun C et al: Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet 2006; 15: 2076–2086.
Acknowledgements
This research was supported by Estonian Science Foundation Grants no. 5574 (to TK) and no. 6040 (to Kristiina Tambets), European Commission project QLG2-CT-2002-90455 (RV); grants of the RAS program ‘Human molecular polymorphism’ (to LZ and EK), the Russian Foundation for Basic Research (Grants 04-04-48639 to LZ and 04-04-48678 to EK), and the RAS program ‘Biodiversity and dynamics of gene pools’ (to LZ and EK).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg).
Supplementary information
Rights and permissions
About this article
Cite this article
Rootsi, S., Zhivotovsky, L., Baldovič, M. et al. A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet 15, 204–211 (2007). https://doi.org/10.1038/sj.ejhg.5201748
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201748
Keywords
This article is cited by
-
Parallel signatures of Mycobacterium tuberculosis and human Y-chromosome phylogeography support the Two Layer model of East Asian population history
Communications Biology (2023)
-
Phylogeographic review of Y chromosome haplogroups in Europe
International Journal of Legal Medicine (2021)
-
Partial-AZFc deletions in Chilean men with primary spermatogenic impairment: gene dosage and Y-chromosome haplogroups
Journal of Assisted Reproduction and Genetics (2020)
-
Relating Clans Ao and Aisin Gioro from northeast China by whole Y-chromosome sequencing
Journal of Human Genetics (2019)
-
Genetic history of Bashkirian Mari and Southern Mansi ethnic groups in the Ural region
Molecular Genetics and Genomics (2019)