Abstract
The Zika virus, which has been implicated in an increase in neonatal microcephaly and Guillain–Barré syndrome, has spread rapidly through tropical regions of the world. The virulence protein NS1 functions in genome replication and host immune-system modulation. Here, we report the crystal structure of full-length Zika virus NS1, revealing an elongated hydrophobic surface for membrane association and a polar surface that varies substantially among flaviviruses.
Similar content being viewed by others
Main
Zika virus (ZIKV) is the cause of a current international public health emergency, owing to its rapid spread in the Americas1 and an anticipated increase in the habitats of the Aedes aegypti mosquito vector. Although ZIKV has been in circulation in tropical regions of the world for more than 60 years2, a causative association with severe congenital microcephaly via placental transmission3,4 and an association with Guillain–Barré syndrome in adults5 had not been identified until the 2015 outbreak in Brazil. The lack of effective vaccines, antiviral treatments and rapid detection methods makes detailed investigations of ZIKV and distinct aspects of its life cycle an urgent priority.
ZIKV is a flavivirus related to dengue virus (DENV), West Nile virus (WNV) and Japanese encephalitis virus6,7. The flavivirus positive-sense RNA genome encodes three structural proteins, which form the virus particle, and seven nonstructural proteins, which perform essential functions in genome replication, polyprotein processing and manipulation of cellular processes to viral advantage. Flavivirus nonstructural protein 1 (NS1), one of only ten viral proteins, is a multifunctional virulence factor8,9. Within infected cells, the glycosylated 48-kDa NS1 exists as a membrane-associated dimer after translocation into the endoplasmic reticulum (ER) lumen, where it is essential for viral genome replication. The replication complex at the ER membrane includes NS1 on the lumenal side, viral transmembrane proteins (NS2a, NS2b, NS4a and NS4b) and viral enzymes (NS3 protease–helicase, NS5 capping enzyme and RNA-dependent RNA polymerase) on the cytoplasmic side. Infected cells also secrete NS1 as a hexameric lipoprotein particle10, which is detected in the serum of infected individuals at levels correlated with disease severity. NS1 also associates with the surfaces of infected cells, where its role is unclear. Crystal structures from our laboratory have established details of the dimeric and hexameric architecture of NS1 from dengue virus serotype 2 (DENV2) and WNV11. Secreted NS1 (sNS1) interacts with complement-system proteins and has several immune-modulatory functions. In an animal model, DENV NS1, in the absence of virus, can lead to vascular leakage, which is typical of severe dengue infection12, possibly by activating macrophages via Toll-like receptor 4 (ref. 13). sNS1 is a component of some DENV candidate vaccines. Structure-based mutagenesis has implied additional unexpected NS1 functions during virus maturation, including interaction with the viral prM and envelope proteins14.
Because molecular studies are lacking, the overall characteristics of the ZIKV infection cycle have been inferred from results for several flaviviruses, particularly the DENV, WNV and Japanese encephalitis virus. Recent cryo-EM characterization of the structures of the mature virus particle15,16 and a crystal structure of the C-terminal half of NS1 (ref. 17) have provided details specific to ZIKV. Here, to gain complete structural details for understanding ZIKV NS1 function, we solved a 1.9-Å-resolution structure of the full-length protein from the original Uganda strain, providing insights into membrane interaction and variability in the protein surfaces.
Flavivirus NS1 encompasses three distinct domains: an N-terminal β-roll, an epitope-rich wing domain and a C-terminal β-ladder11,18. Twelve invariant cysteines form six disulfide bonds per monomer. The fundamental unit is a flat cross-shaped dimer, formed via the intertwined β-roll and end-to-end β-ladders (Fig. 1). On the 'inner' face of the dimer, the β-roll domain and an adjacent 'greasy finger' loop form a hydrophobic surface that is the prime candidate for membrane interaction, because it is adjacent to amino acids implicated in contacts with the viral transmembrane proteins19. The dimer 'outer' face is polar and contains the glycosylation sites. In the NS1 hexamer, three dimers assemble with the glycosylated polar faces pointed outward and the hydrophobic faces pointed inward so that they can interact with lipid molecules in the sNS1 lipoprotein particle.
(a) Ribbon representation of the ZIKV NS1 dimer (Uganda strain MR-766), viewed from the outer face. One subunit is in gray, and the other subunit is colored by domain: blue, β-roll (residues 1–29); yellow, wing domain (residues 30–180); orange, connector subdomain with its greasy finger (residues 159–163); red, β-ladder domain (residues 181–352). The wing flexible loop (yellow sticks on both subunits, circled in one subunit) includes residues 108–129, which are not visible in previous structures. Density was clear for all amino acids in one of two subunits in the crystal and for all but residues 113–119 in the second. Glycosylation sites at Asn130 and Asn207 are indicated with black sticks, and disulfides are indicated with yellow double spheres. (b) NS1 dimer viewed along the β-ladder domain, with the hydrophobic face pointed downward, rotated 90° about the horizontal axis relative to the view in a. (c) Electrostatic surface representation of the ZIKV NS1 dimer outer polar face, viewed as in a. A symmetric dimer was created from the complete monomer. (d) Electrostatic representation viewed as in b, with the inner hydrophobic face and aromatic protrusions pointed downward.
In its protein fold and domain arrangement, ZIKV NS1 (Fig. 1a,b and Supplementary Table 1) is virtually identical to DENV2 and WNV NS1 (ref. 11). The wing-domain positions differ by less than 3° rotation relative to the central β-ladder. Despite this overall similarity, the ZIKV NS1 crystal structure provides important new information about a wing-domain flexible loop (residues 108–129) that is not visible in previous structures.
The hydrophobic surface identified in WNV NS1, including the β-roll and a greasy finger11, is expanded in ZIKV NS1, owing to the stabilization of two regions that are disordered (wing flexible loop) or poorly ordered (residues 27–30) in the previous structures (Fig. 2). Interestingly, these segments lie on the inner hydrophobic face of NS1 and include three highly conserved tryptophan residues as well as the dipeptide 123–124, which is hydrophobic in all NS1 sequences and contains an aromatic residue in more than 90% of NS1 sequences (Supplementary Fig. 1). The aromatic side chains in the flexible loop provide a striking outward expansion of the hydrophobic surface to include the ends of the wing domains (Fig. 2b,c). Together with Trp28 in the β-roll domain and Phe163 in the greasy finger, Trp115, Trp118 and Phe123 protrude from the dimer inner hydrophobic face and form an array of conserved aromatic groups that supply additional anchor points for membrane association, particularly with the interfacial region of the membrane bilayer (Fig. 2c,d).
(a) Ordered 'wing tip' (residues 108–129), shown as green sticks with conserved Trp115, Trp118 and Phe123 in yellow. (b) Surface representation of WNV NS1 (end view, as in Fig. 1b). The structure lacks the wing flexible loop, which was disordered in the crystals. The β-roll (blue) and protruding aromatic 'anchors' (residues 28 and 160–163 in yellow) define a limited hydrophobic surface. The β-ladder domain points toward the reader in the center of the image. (c) Surface representation of ZIKV NS1, showing the outward expansion of the hydrophobic surface with the wing flexible loop (green) and protruding aromatic anchors (residues 28, 115, 118, 123 and 160–163, yellow) in the same orientation and with coloring as in b. (d) Surface representation of the hydrophobic face of the NS1 dimer, colored as in b.
The aromatic amino acids on the ZIKV NS1 wing flexible loop appear in a membrane-interacting position, but evidence suggests that this is not the only loop conformation. Linear epitopes mapped to NS1 occur with high frequency in the flexible loop (http://www.iedb.org/), and a high degree of flexibility has been indicated by the lack of electron density for the loop in previous crystal structures, which include a total of ten views of the WNV and DENV2 NS1 polypeptides. None of the flexible-loop aromatic side chains pack into the interior of the wing domain in any crystal structure; therefore, we conclude that these side chains are involved in interactions with the membrane, with lipids and perhaps also with other proteins, in agreement with the results of recent mutational studies showing an interaction of DENV NS1 Trp115 with the envelope proteins, presumably in the context of virions14.
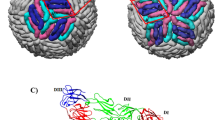
Another aspect of NS1 is its secreted lipoprotein hexameric form. Crystal structures of lipid-free WNV and DENV2 NS1 proteins reveal a hexameric association of three dimers with the hydrophobic surfaces pointed inward11, as expected for a lipid complex. We purified lipid-free recombinant ZIKV NS1 and observed two distinct species: one consistent in size with an NS1 hexamer and the other consistent with a dimer, which yielded the crystal structure reported here. To examine whether ZIKV NS1 forms a hexamer similarly to previously studied flavivirus NS1 proteins, we visualized the hexamer fraction by negative-stain EM (Fig. 3a and Supplementary Fig. 2). Two-dimensional class averages of ZIKV NS1 showed hexameric particles consistent in both size and shape with hexamers of WNV and DENV2 NS1 (refs. 10, 11, 20). The superposition of the ZIKV NS1 dimer on the DENV2 NS1 hexamer structure provided a model for the lipoprotein hexamer of ZIKV NS1 (Fig. 3b,c). In this model, the wing flexible loop is directed inward, toward the lipid cargo. The ordered loop spans a gap between dimers and may potentially stabilize the hexamer. Because the loop residues are not visible in previous crystal structures, it is likely that these segments have different conformations in the context of lipid or protein association of sNS1.
(a) Representative EM class averages of ZIKV NS1 hexamers embedded in negative stain. All 88 class averages are shown in Supplementary Figure 2. (b) Top view of the ZIKV NS1 hexamer, modeled by superposition of three copies of the ZIKV NS1 dimer on dimers within the DENV2 Individual dimers are colored gray, orange and salmon. In the gray NS1 dimer, the wing flexible loop is green (residues 108–129); protruding conserved aromatic side chains (residues 28, 115, 118, 123 and 160–163) are yellow; and the β-roll is blue. (c) Side view of the ZIKV NS1 hexamer model, colored as in b and rotated 90° about the horizontal axis. The salmon-colored dimer is removed to show how the aromatic side chains and hydrophobic β-roll line the interior of the hexamer.
Of the two faces of NS1, the outer polar face exhibits the greater degree of sequence diversity11. This characteristic has implications for immune-system recognition and vaccine development, because the outer face is more likely to elicit an immune response, whether on the cell surface or as a secreted lipoprotein. Even within only the central β-ladder domain, ZIKV NS1, as compared with DENV NS1 and WNV NS1 (ref. 17), exhibits differences in electrostatic surface potential. With the structure of full-length ZIKV NS1, we were able to visualize the surface differences in the context of the complete NS1 proteins, and we further modeled NS1 from an isolate in the Brazil outbreak. ZIKVBrazil NS1 and ZIKVUganda NS1 differ by two nonconservative changes: Glu146Uganda versus Lys146Brazil, and Tyr286 Uganda versus His286 Brazil, as well as seven conservative changes (Supplementary Fig. 1). We also used the ZIKV NS1 structure to model complete DENV2 NS1 and WNV NS1 structures. The complete NS1 proteins have only minor differences in the electrostatic surface potentials of the dimer inner hydrophobic faces, because the protruding β-roll, greasy finger and aromatic expansion (residues 115–123) are hydrophobic (Fig. 1d and Supplementary Fig. 3). In contrast, the surface potentials of the NS1 outer faces vary dramatically among flaviviruses. NS1 proteins from both ZIKV strains display a negatively charged region on the outer face of the wing domain and the central portion of the β-ladder domain, whereas DENV2 NS1 and WNV NS1 are positively charged or neutral in these regions (Supplementary Fig. 3). The sequence changes from ZIKVUganda to ZIKVBrazil illustrate how genetic drift has altered the appearance of the NS1 virulence factor, as viewed by the innate immune system.
The structure of full-length ZIKV NS1 provides a solid molecular framework to allow the multiple functions of this virulence protein to begin to be understood. The structure of the wing flexible loop reveals an expanded surface permitting NS1 to associate with membranes during replication, to associate with immature virions during particle morphogenesis and to facilitate the interactions necessary for formation of the hexameric lipoprotein complex.
Methods
Construction, cloning and expression evaluation.
The amino acid sequence for Zika strain MR766, the original 1947 Ugandan strain, was used to design a gene sequence codon-optimized for insect cell expression. A linear synthetic DNA was purchased from IDT. Cloning, production of baculovirus and small-scale expression evaluation were carried out as described previously21.
Large-scale production and purification of NS1 protein.
Infections at 1 l were carried out in Sf9 cells seeded at 2 × 106 cells/ml and infected with a multiplicity of infection of two. Cells were harvested 72 h after infection. The purification was carried out as previously described for WNV and DENV2 NS1 proteins11.
Crystallization and structure determination.
ZIKV NS1 crystals were grown at 4 °C by vapor-diffusion equilibration of a 1:1 or 1:2 mixture of protein stock (∼5 mg/ml NS1, 50 mM Tris, pH 7.5, 50 mM NH3SO4 and 10% glycerol) and reservoir solution (20–25% PEG 3350, 150 mM NH3SO4 and 100 mM Tris, pH 8.5). Crystals formed within one week, were harvested without additional cryoprotection and were flash cooled in liquid nitrogen. Data were collected at GM/CA beamline 23-ID-D at the Advanced Photon Source at an X-ray energy of 12.0 keV (λ = 1.033 Å) with a Pilatus3 6M detector. A total of 360° of data were collected from a single crystal at 100 K with a 0.2° image width. Data were integrated and scaled with XDS22 (Supplementary Table 1). The crystal structure was solved by molecular replacement, using the WNV NS1 (PDB 4O6D11) as a search model, with Phaser23. The asymmetric unit contained one dimer. Model building was carried out with Coot24, and refinement was carried out with PHENIX25. Electrostatic surface potentials were calculated with APBS26. The model was validated with MolProbity27 and has excellent Ramachandran statistics (97% favored, 3% allowed and 0 outliers).
Negative-stain electron microscopy.
Samples were prepared through the conventional negative-staining protocol28, and imaged at room temperature with a Tecnai T12 electron microscope (FEI Company) operated at 120 kV. 4,122 particle projections were subjected to iterative stable alignment and classification with ISAC29, thus producing 88 classes.
Accession codes.
Coordinates and structure factors for the ZIKV NS1 structure have been deposited in the Protein Data Bank under accession code PDB 5K6K.
References
Gatherer, D. & Kohl, A. J. Gen. Virol. 97, 269–273 (2016).
Dick, G.W.A., Kitchen, S.F. & Haddow, A.J. Trans. R. Soc. Trop. Med. Hyg. 46, 509–520 (1952).
Mlakar, J. et al. N. Engl. J. Med. 374, 951–958 (2016).
Calvet, G. et al. Lancet Infect. Dis. 16, 653–660 (2016).
Cao-Lormeau, V.M. et al. Lancet 387, 1531–1539 (2016).
Lazear, H.M. & Diamond, M.S. J. Virol. 90, 4864–4875 (2016).
Pierson, T.C. & Diamond, M.S. in Fields Virology, Vol. 2 (eds. Knipe, D.M. & Howley, P.) 747–794 (Wolters Kluwer, 2013).
Muller, D.A. & Young, P.R. Antiviral Res. 98, 192–208 (2013).
Watterson, D., Modhiran, N. & Young, P.R. Antiviral Res. 130, 7–18 (2016).
Gutsche, I. et al. Proc. Natl. Acad. Sci. USA 108, 8003–8008 (2011).
Akey, D.L. et al. Science 343, 881–885 (2014).
Beatty, P.R. et al. Sci. Transl. Med. 7, 304ra141 (2015).
Modhiran, N. et al. Sci. Transl. Med. 7, 304ra142 (2015).
Scaturro, P., Cortese, M., Chatel-Chaix, L., Fischl, W. & Bartenschlager, R. PLoS Pathog. 11, e1005277 (2015).
Sirohi, D. et al. Science 352, 467–470 (2016).
Kostyuchenko, V.A. et al. Nature 533, 425–428 (2016).
Song, H., Qi, J., Haywood, J., Shi, Y. & Gao, G.F. Nat. Struct. Mol. Biol. 23, 456–458 (2016).
Edeling, M.A., Diamond, M.S. & Fremont, D.H. Proc. Natl. Acad. Sci. USA 111, 4285–4290 (2014).
Youn, S. et al. J. Virol. 86, 7360–7371 (2012).
Muller, D.A. et al. J. Gen. Virol. 93, 771–779 (2012).
Brown, W.C. et al. Protein Expr. Purif. 77, 34–45 (2011).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A.J. et al. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Davis, I.W. et al. Nucleic Acids Res. 35, W375–W383 (2007).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Biol. Proced. Online 6, 23–34 (2004).
Yang, Z., Fang, J., Chittuluru, J., Asturias, F.J. & Penczek, P.A. Structure 20, 237–247 (2012).
Acknowledgements
The work described in this report was supported by a Margaret J. Hunter Collegiate Professorship in the Life Sciences to J.L.S. and by the University of Michigan Life Sciences Institute. GM/CA@APS is supported by the NIH National Institute of General Medical Sciences (AGM-12006) and the National Cancer Institute (ACB-12002). The Advanced Photon Source is a US Department of Energy (DOE) Office of Science User Facility operated by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
W.C.B., D.L.A., R.J.K. and J.L.S. planned the experiments; W.C.B. created expression constructs; J.R.K. purified and crystallized the protein; D.L.A. solved the crystal structure; J.T.T. and G.S. performed the electron microscopy analysis; W.C.B., D.L.A., R.J.K. and J.L.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Sequence alignment of NS1 proteins.
Sequence alignment of NS1 proteins from ZIKVUganda (MR-766), ZIKVBrazil (KU49755, identical to H/PF/2013), WNV (NY99) and DENV2 (16681). Hydrophobic residues mentioned in the text are highlighted in yellow. Non-conservative differences between ZIKVUganda and ZIKVBrazil are marked with an asterisk and conserved differences with a dot. Colored bars below the alignment represent the β-roll (blue), wing (yellow) with connector sub-domain (orange) and β-ladder (red). The wing domain flexible loop is indicated above the alignment. Every tenth residue is underlined.
Supplementary Figure 2 Negative-stain electron microscopy visualization of the hexamer fraction of ZIKV NS1.
The averages (right) were obtained by classification of 4122 particle projections in raw images such as the representative example at left. Two of the classes from this group are shown in Fig. 3.
Supplementary Figure 3 Variation in surface potential of flavivirus NS1 proteins.
The inner membrane- or lipid-proximal surface (center column) is more hydrophobic (white) than is the outer polar surface (left column). Surfaces are colored by electrostatic potential from −5 kT (red) to +5 kT (blue). On the inner hydrophobic face (right column), the aromatic side chains in the wing flexible loop expand the hydrophobic surface outward from the β-roll at the center of the dimer. For this figure, the NS1 from ZIKVBrazil was modeled from ZIKV NS1Uganda, and the flexible loops were modeled on the DENV2 and WNV NS1 structures based on the ZIKVUganda NS1 structure. The outer polar NS1 surface, which is presented to the host immune proteins, varies greatly among NS1 proteins (left column). Changes in the outer surface are apparent among ZIKV strains by comparison of NS1 proteins from the first-identified 1947 Ugandan strain MR-766, for which the crystal structure is reported (top panel), and from a strain isolated in the 2015 Brazilian outbreak (GenBank KU497555; second panel), which has an identical NS1 sequence to strain H/PF/2013 from the French Polynesian outbreak.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Table 1 (PDF 1372 kb)
Rights and permissions
About this article
Cite this article
Brown, W., Akey, D., Konwerski, J. et al. Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol 23, 865–867 (2016). https://doi.org/10.1038/nsmb.3268
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3268
This article is cited by
-
Compartmentalized replication organelle of flavivirus at the ER and the factors involved
Cellular and Molecular Life Sciences (2021)
-
Antibodies targeting epitopes on the cell-surface form of NS1 protect against Zika virus infection during pregnancy
Nature Communications (2020)
-
Association of high pressure and alkaline condition for solubilization of inclusion bodies and refolding of the NS1 protein from zika virus
BMC Biotechnology (2018)
-
Rewiring cellular networks by members of the Flaviviridae family
Nature Reviews Microbiology (2018)
-
An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction
Nature Communications (2018)