Abstract
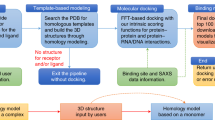
The ClusPro server (https://cluspro.org) is a widely used tool for protein–protein docking. The server provides a simple home page for basic use, requiring only two files in Protein Data Bank (PDB) format. However, ClusPro also offers a number of advanced options to modify the search; these include the removal of unstructured protein regions, application of attraction or repulsion, accounting for pairwise distance restraints, construction of homo-multimers, consideration of small-angle X-ray scattering (SAXS) data, and location of heparin-binding sites. Six different energy functions can be used, depending on the type of protein. Docking with each energy parameter set results in ten models defined by centers of highly populated clusters of low-energy docked structures. This protocol describes the use of the various options, the construction of auxiliary restraints files, the selection of the energy parameters, and the analysis of the results. Although the server is heavily used, runs are generally completed in <4 h.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout













Similar content being viewed by others
References
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574 (2001).
Gavin, A.C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Ho, Y. et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 (2002).
Ewing, R.M. et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89 (2007).
Smith, G.R. & Sternberg, M.J. Prediction of protein-protein interactions by docking methods. Curr. Opin. Struct. Biol. 12, 28–35 (2002).
Halperin, I., Ma, B., Wolfson, H. & Nussinov, R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins 47, 409–443 (2002).
Ritchie, D.W. Recent progress and future directions in protein-protein docking. Curr. Protein Pept. Sci. 9, 1–15 (2008).
Vajda, S. & Kozakov, D. Convergence and combination of methods in protein-protein docking. Curr. Opin. Struct. Biol. 19, 164–170 (2009).
Aloy, P., Ceulemans, H., Stark, A. & Russell, R.B. The relationship between sequence and interaction divergence in proteins. J. Mol. Biol. 332, 989–998 (2003).
Sinha, R., Kundrotas, P.J. & Vakser, I.A. Protein docking by the interface structure similarity: how much structure is needed? PLoS One 7, e31349 (2012).
Comeau, S.R., Gatchell, D.W., Vajda, S. & Camacho, C.J. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32, W96–W99 (2004).
Comeau, S.R., Gatchell, D.W., Vajda, S. & Camacho, C.J. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 (2004).
Comeau, S.R. et al. ClusPro: performance in CAPRI rounds 6-11 and the new server. Proteins 69, 781–785 (2007).
Kozakov, D. et al. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13-19. Proteins 78, 3124–3130 (2010).
Kozakov, D. et al. How good is automated protein docking? Proteins 81, 2159–2166 (2013).
Kozakov, D., Brenke, R., Comeau, S.R. & Vajda, S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 (2006).
Katchalski-Katzir, E. et al. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. USA 89, 2195–2199 (1992).
Gabb, H.A., Jackson, R.M. & Sternberg, M.J.E. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 272, 106–120 (1997).
Mandell, J.G. et al. Protein docking using continuum electrostatics and geometric fit. Protein Eng. 14, 105–113 (2001).
Chen, R. & Weng, Z.P. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins 47, 281–294 (2002).
Kozakov, D., Clodfelter, K.H., Vajda, S. & Camacho, C.J. Optimal clustering for detecting near-native conformations in protein docking. Biophys. J. 89, 867–875 (2005).
Chen, R., Mintseris, J., Janin, J. & Weng, Z. A protein-protein docking benchmark. Proteins 52, 88–91 (2003).
Mintseris, J. et al. Protein-Protein Docking Benchmark 2.0: an update. Proteins 60, 214–216 (2005).
Hwang, H., Pierce, B., Mintseris, J., Janin, J. & Weng, Z. Protein-protein docking benchmark version 3.0. Proteins 73, 705–709 (2008).
Hwang, H., Vreven, T., Janin, J. & Weng, Z. Protein-protein docking benchmark version 4.0. Proteins 78, 3111–3114 (2010).
Vajda, S. & Camacho, C.J. Protein-protein docking: is the glass half-full or half-empty? Trends Biotechnol. 22, 110–116 (2004).
Vajda, S. Classification of protein complexes based on docking difficulty. Proteins 60, 176–180 (2005).
Yershova, A., Jain, S., LaValle, S.M. & Mitchell, J.C. Generating uniform incremental grids on SO(3) using the Hopf Fibration. Int. J. Robot. Res. 29, 801–812 (2010).
Chuang, G.Y., Kozakov, D., Brenke, R., Comeau, S.R. & Vajda, S. DARS (Decoys As the Reference State) potentials for protein-protein docking. Biophys. J. 95, 4217–4227 (2008).
Gilson, M.K. & Honig, B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins 4, 7–18 (1988).
Brooks, B.R. et al. Charmm - a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Lorenzen, S. & Zhang, Y. Identification of near-native structures by clustering protein docking conformations. Proteins 68, 187–194 (2007).
Shortle, D., Simons, K.T. & Baker, D. Clustering of low-energy conformations near the native structures of small proteins. Proc. Natl. Acad. Sci. USA 95, 11158–11162 (1998).
Mendez, R., Leplae, R., Lensink, M.F. & Wodak, S.J. Assessment of CAPRI predictions in rounds 3-5 shows progress in docking procedures. Proteins 60, 150–169 (2005).
Lensink, M.F., Mendez, R. & Wodak, S.J. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins 69, 704–718 (2007).
Lensink, M.F. & Wodak, S.J. Docking and scoring protein interactions: CAPRI 2009. Proteins 78, 3073–3084 (2010).
Lensink, M.F. & Wodak, S.J. Docking, scoring, and affinity prediction in CAPRI. Proteins 81, 2082–2095 (2013).
Lensink, M.F. et al. Prediction of homo- and hetero-protein complexes by protein docking and template-based modeling: a CASP-CAPRI experiment. Proteins 84, 323–348 (2016).
Kamal, J.K., Benchaar, S.A., Takamoto, K., Reisler, E. & Chance, M.R. Three-dimensional structure of cofilin bound to monomeric actin derived by structural mass spectrometry data. Proc. Natl. Acad. Sci. USA 104, 7910–7915 (2007).
Kamal, J.K. & Chance, M.R. Modeling of protein binary complexes using structural mass spectrometry data. Protein Sci. 17, 79–94 (2008).
Luxan, G. et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 19, 193–201 (2013).
Tran, K. et al. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc. Natl. Acad. Sci. USA 111, E738–E747 (2014).
Schwede, T. et al. Outcome of a workshop on applications of protein models in biomedical research. Structure 17, 151–159 (2009).
Sondermann, H., Nagar, B., Bar-Sagi, D. & Kuriyan, J. Computational docking and solution x-ray scattering predict a membrane-interacting role for the histone domain of the Ras activator son of sevenless. Proc. Natl. Acad. Sci. USA 102, 16632–16637 (2005).
Bourdon, A. et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 39, 776–780 (2007).
Cosconati, S., Marinelli, L., Lavecchia, A. & Novellino, E. Characterizing the 1,4-dihydropyridines binding interactions in the L-type Ca2+ channel: model construction and docking calculations. J. Med. Chem. 50, 1504–1513 (2007).
Rumpel, S., Becker, S. & Zweckstetter, M. High-resolution structure determination of the CylR2 homodimer using paramagnetic relaxation enhancement and structure-based prediction of molecular alignment. J. Biomol. NMR 40, 1–13 (2008).
Kucuk, C. et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat. Commun. 6, 6025 (2015).
Ye, Z., Musiol, E.M., Weber, T. & Williams, G.J. Reprogramming acyl carrier protein interactions of an Acyl-CoA promiscuous trans-acyltransferase. Chem. Biol. 21, 636–646 (2014).
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R. & Wolfson, H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 (2005).
Steeland, S. et al. Generation and characterization of small single domain antibodies inhibiting human tumor necrosis factor receptor 1. J. Biol. Chem. 290, 4022–4037 (2015).
Bohnuud, T. et al. A benchmark testing ground for integrating homology modeling and protein docking. Proteins (2016) http://dx.doi.org/10.1002/prot.25063.
Chen, R., Li, L. & Weng, Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52, 80–87 (2003).
Brenke, R. et al. Application of asymmetric statistical potentials to antibody-protein docking. Bioinformatics 28, 2608–2614 (2012).
Comeau, S.R. & Camacho, C.J. Predicting oligomeric assemblies: N-mers a primer. J. Struct. Biol. 150, 233–244 (2005).
Pierce, B., Tong, W. & Weng, Z. M-ZDOCK: a grid-based approach for Cn symmetric multimer docking. Bioinformatics 21, 1472–1478 (2005).
Yang, S. Methods for SAXS-based structure determination of biomolecular complexes. Adv. Mater. 26, 7902–7910 (2014).
Xia, B. et al. Accounting for observed small angle X-ray scattering profile in the protein-protein docking server ClusPro. J. Comput. Chem. 36, 1568–1572 (2015).
Pons, C. et al. Structural characterization of protein-protein complexes by integrating computational docking with small-angle scattering data. J. Mol. Biol. 403, 217–230 (2010).
Schneidman-Duhovny, D., Hammel, M. & Sali, A. Macromolecular docking restrained by a small angle X-ray scattering profile. J. Struct. Biol. 173, 461–471 (2011).
Bernfield, M. et al. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68, 729–777 (1999).
Lindahl, U. Heparan sulfate-protein interactions - A concept for drug design? Thromb. Haemostasis 98, 109–115 (2007).
Fugedi, P. The potential of the molecular diversity of heparin and heparan sulfate for drug development. Mini Rev. Med. Chem. 3, 659–667 (2003).
Esko, J.D. & Lindahl, U. Molecular diversity of heparan sulfate. J. Clin. Invest. 108, 169–173 (2001).
Forster, M. & Mulloy, B. Computational approaches to the identification of heparin-binding sites on the surfaces of proteins. Biochem. Soc. Trans. 34, 431–434 (2006).
Mottarella, S.E. et al. Docking server for the identification of heparin binding sites on proteins. J. Chem. Inf. Model. 54, 2068–2078 (2014).
Gray, J.J. et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299 (2003).
Zacharias, M. ATTRACT: protein-protein docking in CAPRI using a reduced protein model. Proteins 60, 252–256 (2005).
Dominguez, C., Boelens, R. & Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003).
de Vries, S.J., van Dijk, M. & Bonvin, A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 (2010).
Janin, J. et al. CAPRI: a critical assessment of predicted interactions. Proteins 52, 2–9 (2003).
Mendez, R., Leplae, R., De Maria, L. & Wodak, S.J. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins 52, 51–67 (2003).
de Vries, S.J., Schindler, C.E., Chauvot de Beauchene, I. & Zacharias, M. A web interface for easy flexible protein-protein docking with ATTRACT. Biophys. J. 108, 462–465 (2015).
Moal, I.H. & Bates, P.A. SwarmDock and the use of normal modes in protein-protein docking. Int. J. Mol. Sci. 11, 3623–3648 (2010).
Ramirez-Aportela, E., Lopez-Blanco, J.R. & Chacon, P. FRODOCK 2.0: fast protein-protein docking server. Bioinformatics 32, 2386–2388 (2016).
Yu, J. et al. InterEvDock: a docking server to predict the structure of protein-protein interactions using evolutionary information. Nucleic Acids Res. 44, W542–W549 (2016).
Pierce, B.G., Hourai, Y. & Weng, Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One 6, e24657 (2011).
Tovchigrechko, A. & Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–W314 (2006).
Kozakov, D., Schueler-Furman, O. & Vajda, S. Discrimination of near-native structures in protein-protein docking by testing the stability of local minima. Proteins 72, 993–1004 (2008).
Moghadasi, M. et al. The impact of side-chain packing on protein docking refinement. J. Chem. Inf. Model. 55, 872–881 (2015).
Padhorny, D. et al. Protein-protein docking by fast generalized Fourier transforms on 5D rotational manifolds. Proc. Natl. Acad. Sci. USA 113, E4286–E4293 (2016).
Gruschus, J.M., Greene, L.E., Eisenberg, E. & Ferretti, J.A. Experimentally biased model structure of the Hsc70/auxilin complex: substrate transfer and interdomain structural change. Protein Sci. 13, 2029–2044 (2004).
Liu, J., Wang, H., Zuo, Y. & Farmer, S.R. Functional interaction between peroxisome proliferator-activated receptor gamma and beta-catenin. Mol. Cell. Biol. 26, 5827–5837 (2006).
Lee, D.Y. et al. Mutagenesis and modeling of the peroxiredoxin (Prx) complex with the NMR structure of ATP-bound human sulfiredoxin implicate aspartate 187 of Prx I as the catalytic residue in ATP hydrolysis. Biochemistry 45, 15301–15309 (2006).
Watson, A.A. et al. The crystal structure and mutational binding analysis of the extracellular domain of the platelet-activating receptor CLEC-2. J. Biol. Chem. 282, 3165–3172 (2007).
Martin, M.C., Allan, L.A., Mancini, E.J. & Clarke, P.R. The docking interaction of caspase-9 with ERK2 provides a mechanism for the selective inhibitory phosphorylation of caspase-9 at threonine 125. J. Biol. Chem. 283, 3854–3865 (2008).
Liu, C. et al. The SH3-like domain switches its interaction partners to modulate the repression activity of mycobacterial iron-dependent transcription regulator in response to metal ion fluctuations. J. Biol. Chem. 283, 2439–2453 (2008).
Pilpa, R.M. et al. Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J. Biol. Chem. 284, 1166–1176 (2009).
Liang, S. et al. Mapping of a microbial protein domain involved in binding and activation of the TLR2/TLR1 heterodimer. J. Immunol. 182, 2978–2985 (2009).
Cohavi, O., Tobi, D. & Schreiber, G. Docking of antizyme to ornithine decarboxylase and antizyme inhibitor using experimental mutant and double-mutant cycle data. J. Mol. Biol. 390, 503–515 (2009).
Hofmann, W.A. et al. SUMOylation of nuclear actin. J. Cell Biol. 186, 193–200 (2009).
Arthur, C.J. et al. Structure and malonyl CoA-ACP transacylase binding of streptomyces coelicolor fatty acid synthase acyl carrier protein. ACS Chem. Biol. 4, 625–636 (2009).
Nuth, M. & Cowan, J.A. Iron-sulfur cluster biosynthesis: characterization of IscU-IscS complex formation and a structural model for sulfide delivery to the [2Fe-2S] assembly site. J. Biol. Inorg. Chem. 14, 829–839 (2009).
Stauch, B. et al. Model structure of APOBEC3C reveals a binding pocket modulating ribonucleic acid interaction required for encapsidation. Proc. Natl. Acad. Sci. USA 106, 12079–12084 (2009).
Venkatachari, N.J. et al. Human immunodeficiency virus type 1 Vpr: oligomerization is an essential feature for its incorporation into virus particles. Virol. J. 7, 119 (2010).
Fredericks, W.J. et al. The bladder tumor suppressor protein TERE1 (UBIAD1) modulates cell cholesterol: Implications for tumor progression. DNA Cell. Biol. 30, 851–864 (2011).
Lira-Navarrete, E. et al. Structural insights into the mechanism of protein o-fucosylation. PLoS One 6, e25365 (2011).
Li, D. et al. A comprehensive model of the spectrin divalent tetramer binding region deduced using homology modeling and chemical cross-linking of a mini-spectrin. J. Biol. Chem. 285, 29535–29545 (2010).
Zhang, G.F. et al. Ligand-independent antiapoptotic function of estrogen receptor-beta in lung cancer cells. Mol. Endocrinol. 24, 1737–1747 (2010).
Naudin, C. et al. The occluding loop of cathepsin B prevents its effective inhibition by human kininogens. J. Mol. Biol. 400, 1022–1035 (2010).
Guzman, L. et al. Blockade of ethanol-induced potentiation of glycine receptors by a peptide that interferes with Gbetagamma binding. J. Pharmacol. Exp. Ther. 331, 933–939 (2009).
Xi, J. et al. Interaction between the T4 helicase-loading protein (gp59) and the DNA polymerase (gp43): A locking mechanism to delay replication during replisome assembly. Biochemistry 44, 4600–4600 (2005).
Nelson, S.W., Yang, J.S. & Benkovic, S.J. Site-directed mutations of T4 helicase loading protein (gp59) reveal multiple modes of DNA polymerase inhibition and the mechanism of unlocking by gp41 helicase. J. Biol. Chem. 281, 8697–8706 (2006).
Sobrado, P. et al. Identification of the binding region of the [2Fe-2S] ferredoxin in stearoyl-acyl carrier protein desaturase: insight into the catalytic complex and mechanism of action. Biochemistry 45, 4848–4858 (2006).
Kedlaya, R.H., Bhat, K.M., Mitchell, J., Darnell, S.J. & Setaluri, V. TRP1 interacting PDZ-domain protein GIPC forms oligomers and is localized to intracellular vesicles in human melanocytes. Arch. Biochem. Biophys. 454, 160–169 (2006).
Li, D., Tang, H.Y. & Speicher, D.W. A structural model of the erythrocyte spectrin heterodimer initiation site determined using homology modeling and chemical cross-linking. J. Biol. Chem. 283, 1553–1562 (2008).
Krauss, U., Losi, A., Gartner, W., Jaeger, K.E. & Eggert, T. Initial characterization of a blue-light sensing, phototropin-related protein from Pseudomonas putida: a paradigm for an extended LOV construct. Phys. Chem. Chem. Phys. 7, 2804–2811 (2005).
Surolia, I., Reddy, G.B. & Sinha, S. Hierarchy and the mechanism of fibril formation in ADan peptides. J. Neurochem. 99, 537–548 (2006).
Alminaite, A. et al. Oligomerization of hantavirus nucleocapsid protein: analysis of the N-terminal coiled-coil domain. J. Virol. 80, 9073–9081 (2006).
Alminaite, A., Backstrom, V., Vaheri, A. & Plyusnin, A. Oligomerization of hantaviral nucleocapsid protein: charged residues in the N-terminal coiled-coil domain contribute to intermolecular interactions. J. Gen. Virol. 89, 2167–2174 (2008).
Juszczyk, P. et al. Binding epitopes and interaction structure of the neuroprotective protease inhibitor cystatin c with beta-amyloid revealed by proteolytic excision mass spectrometry and molecular docking simulation. J. Med. Chem. 52, 2420–2428 (2009).
Brown, K.A., Dayal, S., Ai, X., Rumbles, G. & King, P.W. Controlled assembly of hydrogenase-CdTe nanocrystal hybrids for solar hydrogen production. J. Am. Chem. Soc. 132, 9672–9680 (2010).
Raab, M., Parthasarathi, L., Treumann, A., Moran, N. & Daxecker, H. Differential binding of ICln in platelets to integrin-derived activating and inhibitory peptides. Biochem. Biophys. Res. Commun. 392, 258–263 (2010).
Nan, R. et al. Zinc binding to the Tyr402 and His402 allotypes of complement factor H: possible implications for age-related macular degeneration. J. Mol. Biol. 408, 714–735 (2011).
Sato, K., Crowley, P.B. & Dennison, C. Transient homodimer interactions studied using the electron self-exchange reaction. J. Biol. Chem. 280, 19281–19288 (2005).
Man, P. et al. Accessibility changes within diphtheria toxin T domain when in the functional molten globule state, as determined using hydrogen/deuterium exchange measurements. FEBS J. 277, 653–662 (2010).
Wang, G.S. et al. Solution structure of the phosphoryl transfer complex between the signal transducing proteins HPr and IIA(Glucose) of the Escherichia coli phosphoenolpyruvate : sugar phosphotransferase system. EMBO J. 19, 5635–5649 (2000).
Leysen, S., Vanderkelen, L., Weeks, S.D., Michiels, C.W. & Strelkov, S.V. Structural basis of bacterial defense against g-type lysozyme-based innate immunity. Cell. Mol. Life Sci. 70, 1113–1122 (2013).
Fiser, A. & Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 (2003).
Viswanath, S., Ravikant, D.V. & Elber, R. DOCK/PIERR: web server for structure prediction of protein-protein complexes. Methods Mol. Biol. 1137, 199–207 (2014).
Fernandez-Recio, J., Totrov, M. & Abagyan, R. ICM-DISCO docking by global energy optimization with fully flexible side-chains. Proteins 52, 113–117 (2003).
Esquivel-Rodriguez, J., Filos-Gonzalez, V., Li, B. & Kihara, D. Pairwise and multimeric protein-protein docking using the LZerD program suite. Methods Mol. Biol. 1137, 209–234 (2014).
Terashi, G. et al. The SKE-DOCK server and human teams based on a combined method of shape complementarity and free energy estimation. Proteins 69, 866–872 (2007).
May, A. & Zacharias, M. Protein-protein docking in CAPRI using ATTRACT to account for global and local flexibility. Proteins 69, 774–780 (2007).
Huang, S.Y. & Zou, X. An iterative knowledge-based scoring function for protein-protein recognition. Proteins 72, 557–579 (2008).
Eisenstein, M., Ben-Shimon, A., Frankenstein, Z. & Kowalsman, N. CAPRI targets T29-T42: proving ground for new docking procedures. Proteins 78, 3174–3181 (2010).
Andrusier, N., Mashiach, E., Nussinov, R. & Wolfson, H.J. Principles of flexible protein-protein docking. Proteins 73, 271–289 (2008).
Shen, Y. Improved flexible refinement of protein docking in CAPRI rounds 22-27. Proteins 81, 2129–2136 (2013).
Heo, L., Lee, H. & Seok, C. GalaxyRefineComplex: refinement of protein-protein complex model structures driven by interface repacking. Sci. Rep. 6, 32153 (2016).
Cheng, T.M., Blundell, T.L. & Fernandez-Recio, J. pyDock: electrostatics and desolvation for effective scoring of rigid-body protein-protein docking. Proteins 68, 503–515 (2007).
Zhou, H.X. & Qin, S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics 23, 2203–2209 (2007).
Acknowledgements
This investigation was supported by grants R35 GM118078 and R01 GM061867 from the National Institute of General Medical Sciences to S.V., and grants DBI 1147082, DBI 1458509, and AF 1527292 from the National Science Foundation to S.V. and D.K.
Author information
Authors and Affiliations
Contributions
D.K., D.R.H., B.X., and D.B. developed the server; D.K., D.R.H., D.P., B.X., and K.A.P. performed experiments; S.V., K.A.P., D.R.H., and C.Y. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The PIPER docking program, used in the ClusPro server, has been licensed by Boston University to Acpharis Inc., and to Schrodinger LLC. The companies sublicense the program for commercial use. However, the ClusPro server is free for academic and governmental research.
Supplementary information
Supplementary Table 1
Performance of ClusPro on Docking Benchmark 4.0. (PDF 99 kb)
Rights and permissions
About this article
Cite this article
Kozakov, D., Hall, D., Xia, B. et al. The ClusPro web server for protein–protein docking. Nat Protoc 12, 255–278 (2017). https://doi.org/10.1038/nprot.2016.169
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.169
This article is cited by
-
Development of a novel multi‑epitope vaccine against the pathogenic human polyomavirus V6/7 using reverse vaccinology
BMC Infectious Diseases (2024)
-
Enhancing explainable SARS-CoV-2 vaccine development leveraging bee colony optimised Bi-LSTM, Bi-GRU models and bioinformatic analysis
Scientific Reports (2024)
-
Development and immunological evaluation of an mRNA-based vaccine targeting Naegleria fowleri for the treatment of primary amoebic meningoencephalitis
Scientific Reports (2024)
-
NDUFS4 regulates cristae remodeling in diabetic kidney disease
Nature Communications (2024)
-
Immunoinformatics approaches in developing a novel multi-epitope chimeric vaccine protective against Saprolegnia parasitica
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.