Abstract
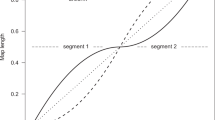
Recombination rates seem to vary extensively along the human genome. Pedigree analysis suggests that rates vary by an order of magnitude when measured at the megabase scale1, and at a finer scale, sperm typing studies point to the existence of recombination hotspots2. These are short regions (1–2 kb) in which recombination rates are 10–1,000 times higher than the background rate. Less is known about how recombination rates change over time. Here we determined to what degree recombination rates are conserved among closely related species by estimating recombination rates from 14 Mb of linkage disequilibrium data in central chimpanzee and human populations. The results suggest that recombination hotspots are not conserved between the two species and that recombination rates in larger (50 kb) genomic regions are only weakly conserved. Therefore, the recombination landscape has changed markedly between the two species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kong, A. et al. A high-resolution recombination map of the human genome. Nat. Genet. 31, 241–247 (2002).
Kauppi, L., Jeffreys, A.J. & Keeney, S. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 5, 413–424 (2004).
Stumpf, M.P. & McVean, G.A. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 4, 959–968 (2003).
Crawford, D.C. et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36, 700–706 (2004).
Fearnhead, P. Consistency of estimators of the population–scaled recombination rate. Theor. Popul. Biol. 64, 67–79 (2003).
Li, N. & Stephens, M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003).
McVean, G.A. et al. The fine-scale structure of recombination rate variation in the human genome. Science 304, 581–584 (2004).
Cullen, M., Perfetto, S.P., Klitz, W., Nelson, G. & Carrington, M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am. J. Hum. Genet. 71, 759–776 (2002).
Kong, A. et al. Recombination rate and reproductive success in humans. Nat. Genet. 36, 1203–1206 (2004).
Wall, J.D., Frisse, L.A., Hudson, R.R. & Di Rienzo, A. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am. J. Hum. Genet. 73, 1330–1340 (2003).
Ptak, S.E. et al. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2, 849–855 (2004).
Hinds, D.A. et al. Whole genome patterns of common DNA variation in diverse human populations. Science (in the press).
Britten, R.J. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc. Natl. Acad. Sci. USA 99, 13633–13635 (2002).
Ebersberger, I., Metzler, D., Schwarz, C. & Paabo, S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70, 1490–1497 (2002).
de Massy, B. Distribution of meiotic recombination sites. Trends Genet. 19, 514–522 (2003).
Petes, T.D. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369 (2001).
Haring, S.J., Halley, G.R., Jones, A.J. & Malone, R.E. Properties of natural double-strand-break sites at a recombination hotspot in Saccharomyces cerevisiae. Genetics 165, 101–114 (2003).
Shiroishi, T., Sagai, T., Hanzawa, N., Gotoh, H. & Moriwaki, K. Genetic control of sex–dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 10, 681–686 (1991).
Jeffreys, A.J. & Neumann, R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat. Genet. 31, 267–271 (2002).
Jeffreys, A.J., Murray, J. & Neumann, R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell 2, 267–273 (1998).
Patil, N. et al. Blocks of limited haplotype diversity revealed by high–resolution scanning of human chromosome 21. Science 294, 1719–1723 (2001).
Altshul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Stephens, M. & Donnelly, P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 (2003).
Stephens, M., Smith, N.J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Dutilleul, P. Modifying the t-test for assessing the correlation between two spatial processes. Biometrics 49, 305–314 (1993).
Hudson, R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002).
Fischer, A., Wiebe, V., Paabo, S. & Przeworski, M. Evidence for a complex demographic history of chimpanzees. Mol. Biol. Evol. 21, 799–808 (2004).
Ptak, S.E., Voelpel, K. & Przeworski, M. Insights into recombination from patterns of linkage disequilibrium in humans. Genetics 167, 387–397 (2004).
Acknowledgements
We thank M. Lachmann, M. Stephens, K. Voepel and J. Wall for computer programs; M. Lachmann, M. Stephens and Rechenzentrum Garching (especially H. Lederer and W. Nagel) for computer support; D. Cox, M. Lachmann, S. Myers, J. Pritchard and M. Stephens for discussions; A. Di Rienzo for comments on the manuscript; N. Shen for her efforts in the hybrid analyses and resequencing array hybridizations; and M. Jen and K. Pant for their contributions to SNP discovery and base calling efforts. P. Morin helped oversee the collection and management of the chimpanzee samples. Support for this work was provided by the Max Planck Society and a Deutsche Forshungsgesellschaft grant to S.P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Comparison of the method and data used here with results from two sperm typing experiments for the MHC region. (PDF 205 kb)
Supplementary Fig. 2
Allele frequency distribution for the chimpanzee and human data. (PDF 88 kb)
Supplementary Table 1
Observed frequency of hotspots. (PDF 66 kb)
Supplementary Table 2
Results from power simulations. (PDF 68 kb)
Supplementary Table 3
Simulation results for other demographic assumptions. (PDF 83 kb)
Rights and permissions
About this article
Cite this article
Ptak, S., Hinds, D., Koehler, K. et al. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet 37, 429–434 (2005). https://doi.org/10.1038/ng1529
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1529
This article is cited by
-
Genetic differentiation and intrinsic genomic features explain variation in recombination hotspots among cocoa tree populations
BMC Genomics (2020)
-
Extensive haplotypes are associated with population differentiation and environmental adaptability in Upland cotton (Gossypium hirsutum)
Theoretical and Applied Genetics (2020)
-
Heterogeneous transposable elements as silencers, enhancers and targets of meiotic recombination
Chromosoma (2019)
-
Characterization of recombination features and the genetic basis in multiple cattle breeds
BMC Genomics (2018)
-
Recombination correlates with synaptonemal complex length and chromatin loop size in bovids—insights into mammalian meiotic chromosomal organization
Chromosoma (2017)