Abstract
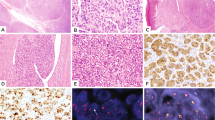
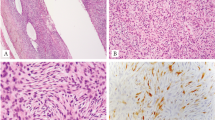
Solitary fibrous tumors (SFTs) are rare mesenchymal tumors. Here, we describe the identification of a NAB2-STAT6 fusion from whole-exome sequencing of 17 SFTs. Analysis in 53 tumors confirmed the presence of 7 variants of this fusion transcript in 29 tumors (55%), representing a lower bound for fusion frequency at this locus and suggesting that the NAB2-STAT6 fusion is a distinct molecular feature of SFTs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Mertens, F. et al. Semin. Oncol. 36, 312–323 (2009).
Svaren, J. et al. Mol. Cell. Biol. 16, 3545–3553 (1996).
Bhattacharyya, S. et al. PLoS ONE 4, e7620 (2009).
Shimoyamada, H. et al. Am. J. Pathol. 177, 70–83 (2010).
Abdulkadir, S.A. et al. Hum. Pathol. 32, 935–939 (2001).
Pal, N.R., Aguan, K., Sharma, A. & Amari, S. BMC Bioinformatics 8, 5 (2007).
Hebenstreit, D., Wirnsberger, G., Horejs-Hoeck, J. & Duschl, A. Cytokine Growth Factor Rev. 17, 173–188 (2006).
Nelson, E.A., Sharma, S.V., Settleman, J. & Frank, D.A. Oncotarget. 2, 518–524 (2011).
Merk, B.C., Owens, J.L., Lopes, M.B., Silva, C.M. & Hussaini, I.M. BMC Cancer 11, 184 (2011).
He, H.J. et al. J. Biol. Chem. 281, 31369–31379 (2006).
Imielinski, M. et al. Cell 150, 1107–1120 (2012).
Hammerman, P.S. et al. Nature 489, 519–525 (2012).
Barbieri, C.E. et al. Nat. Genet. 44, 685–689 (2012).
Pugh, T.J. et al. Nature 488, 106–110 (2012).
Banerji, S. et al. Nature 486, 405–409 (2012).
Acknowledgements
We are grateful to the individuals with SFT who contributed material for this study. We thank members of the Genomics Platform at the Broad Institute for their technical expertise and M. Abazeed and A. Dulak for critically reviewing the manuscript. This work was conducted as part of the Slim Initiative for Genomic Medicine in the Americas (SIGMA), a project funded by the Carlos Slim Health Institute in Mexico. This work was supported by a Specialized Program of Research Excellence (SPORE) grant in Soft Tissue Sarcoma (P50 CA140146) and a P01 grant (P01CA47179) to S. Singer (MSKCC). J.C. is supported by an American Cancer Society AstraZeneca Postdoctoral Fellowship. A.M.C. is supported by a grant from the Kristen Ann Carr Foundation. M.M. is supported by generous donations from M.B. Zuckerman and Team Dragonfly of the Pan-Mass Challenge. Material was collected under institutional review board (IRB)-approved protocols at the Memorial Sloan-Kettering Cancer Center and the Oregon Health & Science University Biolibrary. All patients provided informed consent.
Author information
Authors and Affiliations
Contributions
J.C. and M.M. conceived the project. J.C., A.M.C. and M.M. wrote the manuscript with input from all coauthors. J.C., A.M.C. and R.O. performed experiments. J.C. and M.R. performed computational analyses. A.M. contributed an unpublished algorithm to the analysis. J.C., A.M.C., M.R., S.R.W., R.O., D.A.F. and M.M. analyzed the results. A.M.C. and M.C.H. contributed samples. L.A. and D.A. facilitated the transfer, sequencing and analysis of samples.
Corresponding author
Ethics declarations
Competing interests
M.M. is a paid consultant for and equity holder in Foundation Medicine, a genomics-based oncology diagnostics company, and is a paid consultant for Novartis. M.C.H. is a paid consultant for and equity holder in Molecular MD, a molecular diagnostics company, and is a paid consultant for Novartis.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–4 and Supplementary Tables 1–5 (PDF 2583 kb)
Rights and permissions
About this article
Cite this article
Chmielecki, J., Crago, A., Rosenberg, M. et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet 45, 131–132 (2013). https://doi.org/10.1038/ng.2522
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2522
This article is cited by
-
Complete loss of E-cadherin expression in a rare case of metastatic malignant meningioma: a case report
BMC Neurology (2023)
-
Surgical considerations in the resection of solitary fibrous tumors of the pleura
Journal of Cardiothoracic Surgery (2023)
-
A review of solitary fibrous tumours of the orbit and ocular adnexa
Eye (2023)
-
Postoperative stereotactic radiosurgery for intracranial solitary fibrous tumors: systematic review and pooled quantitative analysis
Journal of Neuro-Oncology (2023)
-
Solitary fibrous tumours involving the genitourinary tract: a case series in rare locations, highlighting the role of STAT6 immunohistochemistry
Virchows Archiv (2023)