Abstract
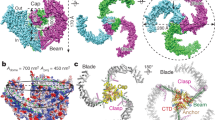
The mechanosensitive Piezo channels function as key eukaryotic mechanotransducers. However, their structures and mechanogating mechanisms remain unknown. Here we determine the three-bladed, propeller-like electron cryo-microscopy structure of mouse Piezo1 and functionally reveal its mechanotransduction components. Despite the lack of sequence repetition, we identify nine repetitive units consisting of four transmembrane helices each—which we term transmembrane helical units (THUs)—which assemble into a highly curved blade-like structure. The last transmembrane helix encloses a hydrophobic pore, followed by three intracellular fenestration sites and side portals that contain pore-property-determining residues. The central region forms a 90?Å-long intracellular beam-like structure, which undergoes a lever-like motion to connect THUs to the pore via the interfaces of the C-terminal domain, the anchor-resembling domain and the outer helix. Deleting extracellular loops in the distal THUs or mutating single residues in the beam impairs the mechanical activation of Piezo1. Overall, Piezo1 possesses a unique 38-transmembrane-helix topology and designated mechanotransduction components, which enable a lever-like mechanogating mechanism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Coste, B. et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010)
Coste, B. et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483, 176–181 (2012)
Ge, J. et al. Architecture of the mammalian mechanosensitive Piezo1 channel. Nature 527, 64–69 (2015)
Zhao, Q. et al. Ion permeation and mechanotransduction mechanisms of mechanosensitive Piezo channels. Neuron 89, 1248–1263 (2016)
Geng, J., Zhao, Q., Zhang, T. & Xiao, B. In touch with the mechanosensitive Piezo channels: structure, ion permeation, and mechanotransduction. Curr. Top. Membr. 79, 159–195 (2017)
Ranade, S. S., Syeda, R. & Patapoutian, A. Mechanically activated ion channels. Neuron 87, 1162–1179 (2015)
Ranade, S. S. et al. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc. Natl Acad. Sci. USA 111, 10347–10352 (2014)
Li, J. et al. Piezo1 integration of vascular architecture with physiological force. Nature 515, 279–282 (2014)
Maksimovic, S. et al. Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature 509, 617–621 (2014)
Woo, S. H. et al. Piezo2 is required for Merkel-cell mechanotransduction. Nature 509, 622–626 (2014)
Ranade, S. S. et al. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516, 121–125 (2014)
Ikeda, R. et al. Merkel cells transduce and encode tactile stimuli to drive Ab-afferent impulses. Cell 157, 664–675 (2014)
Woo, S. H. et al. Piezo2 is the principal mechanotransduction channel for proprioception. Nat. Neurosci. 18, 1756–1762 (2015)
Nonomura, K. et al. Piezo2 senses airway stretch and mediates lung inflation-induced apnoea. Nature 541, 176–181 (2017)
Alper, S. L. Genetic diseases of PIEZO1 and PIEZO2 dysfunction. Curr. Top. Membr. 79, 97–134 (2017)
Kamajaya, A., Kaiser, J. T., Lee, J., Reid, M. & Rees, D. C. The structure of a conserved Piezo channel domain reveals a topologically distinct β sandwich fold. Structure 22, 1520–1527 (2014)
Syeda, R. et al. Piezo1 channels are inherently mechanosensitive. Cell Rep. 17, 1739–1746 (2016)
Cox, C. D. et al. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat. Commun. 7, 10366 (2016)
Wang, Y. & Xiao, B. The mechanosensitive Piezo1 channel: structural features and molecular bases underlying its ion permeation and mechanotransduction. J. Physiol. (Lond.) (2017)
Coste, B. et al. Piezo1 ion channel pore properties are dictated by C-terminal region. Nat. Commun. 6, 7223 (2015)
Prole, D. L. & Taylor, C. W. Identification and analysis of putative homologues of mechanosensitive channels in pathogenic protozoa. PLoS One 8, e66068 (2013)
Zhang, T., Chi, S., Jiang, F., Zhao, Q. & Xiao, B. A protein interaction mechanism for suppressing the mechanosensitive Piezo channels. Nat. Commun. 8, 1797 (2017)
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012)
Gnanasambandam, R., Bae, C., Gottlieb, P. A. & Sachs, F. Ionic selectivity and permeation properties of human PIEZO1 channels. PLoS ONE 10, e0125503 (2015)
Lukacs, V. et al. Impaired PIEZO1 function in patients with a novel autosomal recessive congenital lymphatic dysplasia. Nat. Commun. 6, 8329 (2015)
Syeda, R. et al. Chemical activation of the mechanotransduction channel Piezo1. eLife 4, (2015)
Chang, G., Spencer, R. H., Lee, A. T., Barclay, M. T. & Rees, D. C. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science 282, 2220–2226 (1998)
Brohawn, S. G., Campbell, E. B. & MacKinnon, R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516, 126–130 (2014)
Jin, P. et al. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122 (2017)
Li, X., Zheng, S., Agard, D. A. & Cheng, Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192, 174–178 (2015)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Shaikh, T. R. et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protocols 3, 1941–1974 (2008)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Gong, X. et al. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell 165, 1467–1478 (2016)
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015)
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 184, 226–236 (2013)
Tusnády, G. E. & Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 (2001)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012)
Acknowledgements
We thank N. Yan for critical discussion, and the Beijing Advanced Innovation Center for Structural Biology for facility and financial support. This work was supported by grant numbers 31630090, 2016YFA0500402, 31422027, 31371118 and 2015CB910102 to B.X.; 31570730, 2016YFA0501102 and 2016YFA0501902 to X.L.; and 21375010 to M.-Q.D., from either the National Natural Science Foundation of China or the National Key R&D Program of China. B.X. and X.L. are awardees of the Young Thousand Talent Program of China.
Author information
Authors and Affiliations
Contributions
Q.Z. performed cloning, protein purification, immunostaining and EM data collection; H.Z. performed EM sample preparation, data collection and analysis; S.C., Y.W., J.G., K.W., W.L. and T.Z. carried out biochemical and functional studies; J.W. performed mass spectrometry under the supervision of M.-Q.D.; J.W. built the model; X.L. directed the EM data collection and analysis; B.X. conceived and directed the study, analysed the structure and assisted in model building, made figures and wrote the manuscript with help from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks E. McCleskey and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Purification and cryo-EM analysis of Piezo1.
a, A representative trace of gel filtration of the full-length mouse Piezo1. UV, ultraviolet. The experiment was independently repeated more than three times with similar results. b, A representative cryo-electron micrograph of Piezo1. The experiment was independently repeated more than three times with similar results. c, Power spectrum of the micrograph in b, with the 2.74?Å frequency indicated. d, Representative 2D class averages of Piezo1 particles. e, Euler angle distribution of particles used in the final 3D reconstruction, the height of the cylinder is proportional to the number of particles for that view. f, Gold-standard Fourier shell correlation (FSC) curves of the final density map. The FSC curves were calculated with (purple) or without (red) the application of a soft mask to the two half-set maps. The final FSC curve (blue) was corrected for the soft-mask-induced effect. Reported resolutions were based on the FSC?=?0.143 criteria. g, The final 3D density map of Piezo1 shown in the indicated views is coloured according to the local resolutions estimated by the software Blocres.
Extended Data Figure 2 Flowchart of EM data processing.
Details of data processing are described in the ‘Image processing’ section of the Methods.
Extended Data Figure 3 Subtraction of the projection.
a, Subtraction of the projection of the cap and the other two blades. A distinguishable map of THU4, comprising TM13–TM16, is shown in the red dashed box. Intracellular helical layer, containing several α-helices respectively connecting to TM29, TM25, TM21, TM17 and TM13, is highlighted in the black dashed box. b, Subtraction of the projection of the three blades projection. Identifiable linkers between OH and CED as well as IH and CED are shown in the red dashed box.
Extended Data Figure 4 View of the indicated structural domains illustrates the quality of the cryo-EM density of Piezo1.
The helices are shown in cartoon representation with side chains as sticks. The cryo-EM density is shown as grey mesh.
Extended Data Figure 5 Membrane topology of mouse Piezo1.
On the basis of various membrane topology prediction algorithms, the N-terminal region contains unanimously predicted THUs (highlighted in the purple boxes) that show typical features of the structurally revealed THU7–THU9. On the basis of the resolved 3D structure and the predicted THUs, we propose that mouse Piezo1 possesses a 38-TM topology comprising 9 tandem THUs and the OH and IH (top panel). Diagrams were drawn using the TOPO2 program.
Extended Data Figure 6 The TM27–TM28 loop containing the S1240 and D1260 residues is located at the extracellular side.
Immunofluorescent staining images of cells transfected with the indicated constructs using the anti-Flag antibody either in live-labelling (top) or after fixation and permeabilization (bottom). Scale bars, 10?μm. GFP, green fluorescent protein; IRES, internal ribozyme entry site. The experiments were repeated in two coverslips with similar results.
Extended Data Figure 7 Chemically cross-linked lysine–lysine and lysine–cysteine pairs identified in mouse Piezo1.
a, b, Purified mouse Piezo1 proteins were cross-linked with BS3/DSS (a) or sulfo-GMBS (b), and then digested with trypsin. Following LC–MS/MS analysis of the peptides, cross-linked lysine pairs were identified using pLink. c, The diagram shows the cross-linked lysine pairs between residues located in the beam and other regions. The atom-to-atom distance of the cross-linked residues is shown.
Extended Data Figure 8 Conformational heterogeneity of Piezo1.
Nine classes of Piezo1 structures resulting from symmetry-free 3D classification. Conformational heterogeneity is shown by comparing different classes with the low-passed 6?Å map of the 3.97?Å map with C3 symmetry. Red arrows represent the relative movements of the blades and beams.
Extended Data Figure 9 Characterization of the Piezo1 deletion mutants.
a, b, Immunofluorescent staining images of cells transfected with the indicated constructs using the anti-Flag antibody either in live-labelling (top) or after fixation and permeabilization (bottom). Scale bars, 10?μm. The experiments were repeated in two coverslips with similar results. c, Cell surface biotinylation assay showing comparable plasma membrane expression of the indicated constructs. The experiment was independently repeated for two times with similar results. d, The glutathione S-transferase (GST)-tagged proteins were pulled-down by glutathione beads, followed with western blotting using the anti-GST antibody. The experiment was independently repeated twice with similar results. e, Scatter plot of the maximal poking-induced currents of Piezo1-knockout HEK293 cells transfected with the indicated mutants, which were normalized to the mouse Piezo1 current. f, Scatter plot of the maximal poking-induced currents of HEK293T cells transfected with the indicated constructs in the presence of 30?μM Yoda1. ***P?<?0.0001, one-way ANOVA with Dunn’s multiple comparison test. g, h, Scatter plot of the inactivation tau of HEK293T cells transfected with the indicated constructs in the absence (g) or presence (h) of 30?μM Yoda1. In g, P?=?0.2267 and 0.4177 (for ΔL15–16 and ΔL19–20, respectively) and in h, P?=?0.6263 and 0.6934 (for ΔL15–16 and ΔL19–20, respectively); one-way ANOVA with Dunn’s multiple comparison test (compared with Piezo1). i, Representative traces of the fluorescence signal change of the genetically encoded Ca2+ indicator, GCAMP6s, from HEK293T cells co-transfected with the indicated constructs and GCAMP6s, in response to 30?μM Yoda1. The experiment was independently repeated three times with similar results. Data in e–h are mean?±?s.e.m., and the numbers of recorded cells are indicated above the bars.
Extended Data Figure 10 Characterization of the L1342A/L1345A mutant.
a, Immunofluorescent staining images with the anti-Flag antibody either in live-labelling (top) or after fixation and permeabilization (bottom). The experiment was independently repeated three times with similar results. b, Cell-surface biotinylation assay showing comparable plasma membrane expression of the indicated constructs. The experiment was independently repeated twice with similar results. c, Representative single-channel current traces of the indicated constructs recorded at −140?mV. d, Linear regression fit of average I–V relationships of single-channel recordings of the indicated constructs. The number of recorded cells for Piezo1 and the L1342A/L1345A mutant is 5 and 7, respectively. e, Scatter plot of the unitary conductance calculated from fit of individual recordings. P?=?0.2541, unpaired, two tailed Student’s t-test. f, Scatter plot of the Imax of poking-induced currents. P?=?0.8076, unpaired, two tailed Student’s t-test. g, Structural representation of the L1342 and L1345 residues and those residues in close proximity. h, Scatter plot of the Imax of poking-induced currents of cells transfected with the indicated mutants. P?=?0.9976, 0.9281, 0.3804 and 0.9997 for T2103A, R2104A, L2512A and T2516A, respectively; one-way ANOVA with Dunn’s multiple comparison test. Data in d–f and h are mean?±?s.e.m., and the numbers of recorded cells in e and h are indicated above the bars.
Supplementary information
Supplementary Information
This file contains Supplementary Data Figures 1-2 and Supplementary Table 1. (PDF 4978 kb)
Rights and permissions
About this article
Cite this article
Zhao, Q., Zhou, H., Chi, S. et al. Structure and mechanogating mechanism of the Piezo1 channel. Nature 554, 487–492 (2018). https://doi.org/10.1038/nature25743
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25743
This article is cited by
-
Effects of immediate loading directionality on the mechanical sensing protein PIEZO1 expression and early-stage healing process of peri-implant bone
BioMedical Engineering OnLine (2024)
-
Piezo1 channel exaggerates ferroptosis of nucleus pulposus cells by mediating mechanical stress-induced iron influx
Bone Research (2024)
-
PIEZO1 loss-of-function compound heterozygous mutations in the rare congenital human disorder Prune Belly Syndrome
Nature Communications (2024)
-
Piezo1, the new actor in cell volume regulation
Pflügers Archiv - European Journal of Physiology (2024)
-
Piezo2 Contributes to Traumatic Brain Injury by Activating the RhoA/ROCK1 Pathways
Molecular Neurobiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.