Abstract
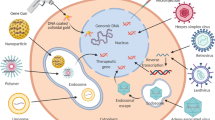
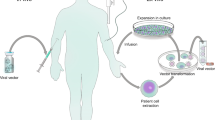
Recent clinical trials of gene therapy have shown remarkable therapeutic benefits and an excellent safety record. They provide evidence for the long-sought promise of gene therapy to deliver 'cures' for some otherwise terminal or severely disabling conditions. Behind these advances lie improved vector designs that enable the safe delivery of therapeutic genes to specific cells. Technologies for editing genes and correcting inherited mutations, the engagement of stem cells to regenerate tissues and the effective exploitation of powerful immune responses to fight cancer are also contributing to the revitalization of gene therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Naldini, L. Ex vivo gene transfer and correction for cell-based therapies. Nature Rev. Genet. 12, 301–315 (2011).
Hacein-Bey-Abina, S. et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 363, 355–364 (2010).
Aiuti, A. et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 360, 447–458 (2009).
Ferrua, F., Brigida, I. & Aiuti, A. Update on gene therapy for adenosine deaminase-deficient severe combined immunodeficiency. Curr. Opin. Allergy Clin. Immunol. 10, 551–556 (2010).
Fischer, A., Hacein- Bey-Abina, S. & Cavazzana-Calvo, M. 20 years of gene therapy for SCID. Nature Immunol. 11, 457–460 (2010). A comprehensive review of the therapeutic potential, risks and limitations of HSC-based SCID gene therapy using γ-RV by some of its pioneers; see also refs 3 and 7.
Gaspar, H. B. et al. Hematopoietic stem cell gene therapy for adenosine deaminase-deficient severe combined immunodeficiency leads to long-term immunological recovery and metabolic correction. Sci. Transl. Med. 3, 97ra80 (2011); erratum 5, 168er1 (2013).
Gaspar, H. B. et al. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 3, 97ra79 (2011).
Boztug, K. et al. Stem-cell gene therapy for the Wiskott―Aldrich syndrome. N. Engl. J. Med. 363, 1918–1927 (2010).
Candotti, F. et al. Gene therapy for adenosine deaminase-deficient severe combined immune deficiency: clinical comparison of retroviral vectors and treatment plans. Blood 120, 3635–3646 (2012).
Kang, E. M. et al. Retrovirus gene therapy for X-linked chronic granulomatous disease can achieve stable long-term correction of oxidase activity in peripheral blood neutrophils. Blood 115, 783–791 (2010).
Hacein-Bey-Abina, S. et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118, 3132–3142 (2008).
Howe, S. J. et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118, 3143–3150 (2008).
Stein, S. et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nature Med. 16, 198–204 (2010).
Braun, C. J. et al. Gene therapy for Wiskott–Aldrich syndrome―long-term efficacy and genotoxicity. Sci. Transl. Med. 6, 227ra233 (2014).
Kang, H. J. et al. Retroviral gene therapy for X-linked chronic granulomatous disease: results from phase I/II trial. Mol. Ther. 19, 2092–2101 (2011).
Aiuti, A. et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 341, 1233151 (2013). In this study, vector insertional analyses in patients show data consistent with improved safety of lentiviral vectors versus γ-RVs while achieving similarly effective disease correction; see also ref. 17.
Hacein-Bey Abina, S. et al. Outcomes following gene therapy in patients with severe Wiskott–Aldrich syndrome. J. Am. Med. Assoc. 313, 1550–1563 (2015).
Hacein-Bey-Abina, S. et al. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 371, 1407–1417 (2014).
Cavazzana-Calvo, M. et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 467, 318–322 (2010).
Cartier, N. et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326, 818–823 (2009). The first trial of HSC gene therapy performed with lentiviral vectors shows data consistent with stable HSC transduction, with long-term safety and efficacy revealed in the follow-up paper (see ref. 21).
Cartier, N. et al. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol. 507, 187–198 (2012).
Biffi, A. et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341, 1233158 (2013). This study highlights the potential of genetic engineering by achieving the stable reconstitution of haematopoiesis in which up to 90% of cells are gene corrected and overexpress the transgene, which provides therapeutic benefit when conventional HSC transplantation is less satisfactory.
Notarangelo, L. D. et al. Primary immunodeficiencies: 2009 update. J. Allergy Clin. Immunol. 124, 1161–1178 (2009).
Kemp, S., Berger, J. & Aubourg, P. X-linked adrenoleukodystrophy: clinical, metabolic, genetic and pathophysiological aspects. Biochim. Biophys. Acta 1822, 1465–1474 (2012).
Gieselmann, V. & Krageloh-Mann, I. Metachromatic leukodystrophy—an update. Neuropediatrics 41, 1–6 (2010).
Gennery, A. R. et al. Transplantation of hematopoietic stem cells and long-term survival for primary immunodeficiencies in Europe: entering a new century, do we do better? J. Allergy Clin. Immunol. 126, 602–610 (2010).
Krägeloh-Mann, I. et al. Juvenile metachromatic leukodystrophy 10 years post transplant compared with a non-transplanted cohort. Bone Marrow Transplant. 48, 369–375 (2013).
Copelan, E. A. Hematopoietic stem-cell transplantation. N. Engl. J. Med. 354, 1813–1826 (2006).
Biffi, A. et al. Correction of metachromatic leukodystrophy in the mouse model by transplantation of genetically modified hematopoietic stem cells. J. Clin. Invest. 113, 1118–1129 (2004).
Biffi, A. et al. Gene therapy of metachromatic leukodystrophy reverses neurological damage and deficits in mice. J. Clin. Invest. 116, 3070–3082 (2006).
Capotondo, A. et al. Brain conditioning is instrumental for successful microglia reconstitution following hematopoietic stem cell transplantation. Proc. Natl Acad. Sci. USA 109, 15018–15023 (2012).
Mingozzi, F. & High, K. A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nature Rev. Genet. 12, 341–355 (2011).
Nayak, S. & Herzog, R. W. Progress and prospects: immune responses to viral vectors. Gene Ther. 17, 295–304 (2010).
Mingozzi, F. & High, K. A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood 122, 23–36 (2013).
Grieger, J. C. & Samulski, R. J. Adeno-associated virus vectorology, manufacturing, and clinical applications. Methods Enzymol. 507, 229–254 (2012).
High, K. H., Nathwani, A., Spencer, T. & Lillicrap, D. Current status of haemophilia gene therapy. Haemophilia 20 (suppl. 4), 43–49 (2014).
Berntorp, E. & Shapiro, A. D. Modern haemophilia care. Lancet 379, 1447–1456 (2012).
Manno, C. S. et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nature Med. 12, 342–347 (2006); erratum 12, 592 (2006). The first clinical data to show the safety and potential efficacy of liver-directed AAV gene transfer, which was unexpectedly abrogated by an immune response against viral capsids (as detailed in ref. 39).
Mingozzi, F. et al. CD8+ T-cell responses to adeno-associated virus capsid in humans. Nature Med. 13, 419–422 (2007).
Nathwani, A. C. et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 371, 1994–2004 (2014). This AAV8-based trial was first to report stable FIX expression at therapeutic levels and also first to overcome the detrimental effect of the immune response to viral capsids by corticosteroid administration.
Rosenberg, S. A. & Restifo, N. P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). A comprehensive review of the clinical development and potentially transformative impact of adoptive T-cell therapy on cancer by one of its pioneers; see also ref. 42.
Maus, M. V. et al. Adoptive immunotherapy for cancer or viruses. Annu. Rev. Immunol. 32, 189–225 (2014).
Dudley, M. E. et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 298, 850–854 (2002).
Gubin, M. M. et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014).
Yadav, M. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). A timely review on the origin and nature of tumour neoantigens and how they can be identified and potentially exploited for targeted T-cell gene therapy in the clinical setting.
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Hunder, N. N. et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N. Engl. J. Med. 358, 2698–2703 (2008).
Johnson, L. A. et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114, 535–546 (2009).
Robbins, P. F. et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 21, 1019–1027 (2015).
Kochenderfer, J. N. et al. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol. 33, 540–549 (2015).
Brentjens, R. J. et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 5, 177ra38 (2013).
Maude, S. L. et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014).
Lee, D. W. et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528 (2015).
June, C. H., Riddell, S. R. & Schumacher, T. N. Adoptive cellular therapy: a race to the finish line. Sci. Transl. Med. 7, 280ps7 (2015).
Biffi, A. et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood 117, 5332–5339 (2011).
Deichmann, A. et al. Insertion sites in engrafted cells cluster within a limited repertoire of genomic areas after gammaretroviral vector gene therapy. Mol. Ther. 19, 2031–2039 (2011).
Doulatov, S., Notta, F., Laurenti, E. & Dick, J. E. Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–136 (2012).
Gattinoni, L. Memory T cells officially join the stem cell club. Immunity 41, 7–9 (2014).
Cieri, N. et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood 121, 573–584 (2013).
Biasco, L. et al. In vivo tracking of T cells in humans unveils decade-long survival and activity of genetically modified T memory stem cells. Sci. Transl. Med. 7, 273ra13 (2015).
Asokan, A., Schaffer, D. V. & Samulski, R. J. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 20, 699–708 (2012).
Mingozzi, F. et al. Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci. Transl. Med. 5, 194ra92 (2013).
Lisowski, L. et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 506, 382–386 (2014).
Kohn, D. B. Gene therapy outpaces haplo for SCID-X1. Blood 125, 3521–3522 (2015).
Logan, A. C., Weissman, I. L. & Shizuru, J. A. The road to purified hematopoietic stem cell transplants is paved with antibodies. Curr. Opin. Immunol. 24, 640–648 (2012).
Provasi, E. et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nature Med. 18, 807–815 (2012).
Torikai, H. et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood 119, 5697–5705 (2012).
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61 (2015).
Li, H. et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood 117, 3311–3319 (2011).
Chandler, R. J. et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J. Clin. Invest. 125, 870–880 (2015).
Nault, J.-C. et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nature Genet. http://dx.doi.org/10.1038/ng.3389 (2015).
Martino, A. T. et al. Engineered AAV vector minimizes in vivo targeting of transduced hepatocytes by capsid-specific CD8+ T cells. Blood 121, 2224–2233 (2013).
Kotterman, M. A. & Schaffer, D. V. Engineering adeno-associated viruses for clinical gene therapy. Nature Rev. Genet. 15, 445–451 (2014).
Cantore, A. et al. Liver-directed lentiviral gene therapy in a dog model of hemophilia B. Sci. Transl. Med. 7, 277ra28 (2015).
Cox, D. B., Platt, R. J. & Zhang, F. Therapeutic genome editing: prospects and challenges. Nature Med. 21, 121–131 (2015).
Tebas, P. et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 370, 901–910 (2014). The first clinical testing of targeted gene disruption that showed the safety, persistence and survival advantage of T cells that have been genetically edited for resistance to HIV-1.
Li, L. et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol. Ther. 21, 1259–1269 (2013).
Bauer, D. E. et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–257 (2013).
Lombardo, A. et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nature Methods 8, 861–869 (2011).
Rio, P. et al. Targeted gene therapy and cell reprogramming in Fanconi anemia. EMBO Mol. Med. 6, 835–848 (2014).
Genovese, P. et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240 (2014). This paper demonstrates differential permissiveness to targeted genome editing in haematopoietic stem and progenitor cells and provides a proof of concept for the in situ correction of SCID-X1 mutations in HSCs.
Hoban, M. D. et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, 2597–2604 (2015).
Osborn, M. J. et al. Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum. Gene Ther. 26, 114–126 (2015).
Jasin, M. & Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, a012740 (2013).
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). In this review, the researchers who pioneered the application of RNA-guided nucleases to genome engineering show how this transformative technique can make targeted genome editing easy.
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature Biotechnol. 29, 816–823 (2011).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature Biotechnol. 33, 187–197 (2015).
Beane, J. D. et al. Clinical scale zinc finger nuclease-mediated gene editing of PD-1 in tumor infiltrating lymphocytes for the treatment of metastatic melanoma. Mol. Ther. 23, 1380–1390 (2015).
Li, H. et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221 (2011). The first study to show the feasibility of targeted genome editing in vivo by the AAV-mediated delivery of artificial nucleases and template.
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nature Biotechnol. 32, 551–553 (2014).
Barzel, A. et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 517, 360–364 (2015).
Simonelli, F. et al. Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 18, 643–650 (2010).
Jacobson, S. G. et al. Improvement and decline in vision with gene therapy in childhood blindness. N. Engl. J. Med. 372, 1920–1926 (2015).
Bainbridge, J. W. et al. Long-term effect of gene therapy on Leber's congenital amaurosis. N. Engl. J. Med. 372, 1887–1897 (2015).
Testa, F. et al. Three-year follow-up after unilateral subretinal delivery of adeno-associated virus in patients with Leber congenital amaurosis type 2. Ophthalmology 120, 1283–1291 (2013).
Wright, A. F. Long-term effects of retinal gene therapy in childhood blindness. N. Engl. J. Med. 372, 1954–1955 (2015).
Leone, P. et al. Long-term follow-up after gene therapy for canavan disease. Sci. Transl. Med. 4, 165ra163 (2012).
Tardieu, M. et al. Intracerebral administration of adeno-associated viral vector serotype rh.10 carrying human SGSH and SUMF1 cDNAs in children with mucopolysaccharidosis type IIIA disease: results of a phase I/II trial. Hum. Gene Ther. 25, 506–516 (2014).
Palfi, S. et al. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson's disease: a dose escalation, open-label, phase 1/2 trial. Lancet 383, 1138–1146 (2014).
Miest, T. S. & Cattaneo, R. New viruses for cancer therapy: meeting clinical needs. Nature Rev. Microbiol. 12, 23–34 (2014).
Lichty, B. D., Breitbach, C. J., Stojdl, D. F. & Bell, J. C. Going viral with cancer immunotherapy. Nature Rev. Cancer 14, 559–567 (2014).
Ogwang, C. et al. Prime-boost vaccination with chimpanzee adenovirus and modified vaccinia Ankara encoding TRAP provides partial protection against Plasmodium falciparum infection in Kenyan adults. Sci. Transl. Med. 7, 286re5 (2015).
Rampling, T. et al. A monovalent chimpanzee adenovirus ebola vaccine — preliminary report. N. Engl. J. Med. http://dx.doi.org/10.1056/NEJMoa1411627 (2015).
Balazs, A. B. et al. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nature Med. 20, 296–300 (2014).
Girard-Gagnepain, A. et al. Baboon envelope pseudotyped LVs outperform VSV-G-LVs for gene transfer into early-cytokine-stimulated and resting HSCs. Blood 124, 1221–1231 (2014).
Baltimore, D. et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science 348, 36–38 (2015).
Bosley, K. S. et al. CRISPR germline engineering—the community speaks. Nature Biotechnol. 33, 478–486 (2015).
Baxter. Baxalta reports continued progress on phase 1/2 clinical trial of BAX335, investigational gene therapy treatment for hemophilia B. Baxter http://www.baxter.com/news-media/newsroom/press-releases/2015/06_24_15_bax335.page (2015).
Montini, E. et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nature Biotechnol. 24, 687–696 (2006).
Modlich, U. et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood 108, 2545–2553 (2006).
Zychlinski, D. et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 16, 718–725 (2008).
Montini, E. et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 119, 964–975 (2009). This preclinical study highlights important features of vector design that affect genotoxicity and reveals strategies to alleviate it; the study was instrumental in promoting the clinical testing of improved vectors (see refs 112–113 for an in vitro assay that provides complementary information).
Modlich, U. et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol. Ther. 17, 1919–1928 (2009).
Zhou, S. et al. A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood 116, 900–908 (2010).
Zhou, S. et al. Mouse transplant models for evaluating the oncogenic risk of a self-inactivating XSCID lentiviral vector. PLoS ONE 8, e62333 (2013).
Baum, C., Modlich, U., Gohring, G. & Schlegelberger, B. Concise review: managing genotoxicity in the therapeutic modification of stem cells. Stem Cells 29, 1479–1484 (2011).
Amendola, M., Venneri, M. A., Biffi, A., Vigna, E. & Naldini, L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nature Biotechnol. 23, 108–116 (2005).
Greco, R. et al. Improving the safety of cell therapy with the TK-suicide gene. Front. Pharmacol. 6, 95 (2015).
Melchiorri, D. et al. Regulatory evaluation of Glybera in Europe — two committees, one mission. Nature Rev. Drug Discov. 12, 719 (2013).
Morrison, C. $1-million price tag set for Glybera gene therapy. Nature Biotechnol. 33, 217–218 (2015).
Brennan, T. A. & Wilson, J. M. The special case of gene therapy pricing. Nature Biotechnol. 32, 874–876 (2014).
Acknowledgements
L.N. apologizes to the many scientists whose contributions to the field could not be acknowledged owing to space limitations. He thanks past and present members of his laboratory and colleagues at the San Raffaele Telethon Institute for Gene Therapy (TIGET) and the San Raffaele Scientific Institute. L.N. is also grateful to the Telethon Foundation, the European Union (FP7 and ERC), the Italian Association for Cancer Research, and the Italian ministries of health and of scientific research for supporting his research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
L.N. is listed as an inventor on pending and issued patents on lentiviral-vector technology, microRNA-regulated vectors and targeted genome editing that were filed by the Salk Institute for Biological Studies, Cell Genesys, the Telethon Foundation and the San Raffaele Scientific Institute. As the director of TIGET, L.N. is involved in a strategic alliance for the development (to marketing authorization) of HSC-based gene therapies for some rare diseases with GlaxoSmithKline (GSK), which licensed metachromatic leukodystrophy and WAS gene therapies in 2014 and became the financial sponsor of the trials. L.N. has established research collaborations on targeted genome editing in HSCs with Sangamo BioSciences and on lentiviral gene therapy of haemophilia with Biogen. L.N. is a founder, owns equity in and chairs the scientific advisory board of Genenta Science, a biotechnology start-up company that aims to develop gene therapy for tumours using tumour-infiltrating monocytes.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Naldini, L. Gene therapy returns to centre stage. Nature 526, 351–360 (2015). https://doi.org/10.1038/nature15818
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15818
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.