Abstract
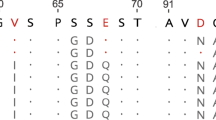
The convergent evolution of a fermentative foregut in two groups of mammals offers an opportunity to study adaptive evolution at the protein level. The appearance of this mode of digestion has been accompanied by the recruitment of lysozyme as a bacteriolytic enzyme in the stomach both in the ruminants (for example the cow) and later in the colobine monkeys (for example the langur). The stomach lysozymes of these two groups share some physicochemical and catalytic properties that appear to adapt them for functioning in the stomach fluid1,2. To examine the basis for these shared properties, we sequenced langur stomach lysozyme and compared it to other lysozymes of known sequence. Tree analysis suggests that, after foregut fermentation arose in monkeys, the langur lysozyme gained sequence similarity to cow stomach lysozyme and evolved two times faster than the other primate lysozymes. This rapid evolution, coupled with functional and sequence convergence upon cow stomach lysozyme, could imply that positive darwinian selection has driven about 50% of the evolution of langur stomach lysozyme.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
1. Dobson, D. E., Prager, E. M. & Wilson, A. C. /. biol. Chem. 259, 11607-11616 (1984). 2. Stewart, C.-B. R. thesis, Univ. Calif., Berkeley (1986). 3. Kimura, M. The Neutral Theory of Molecular Evolution (Cambridge University Press, Cambridge, 1983). 4. Perutz, M. F. Molec. Biol EvoL 1, 1-28 (1983). 5. Hill, R. E. & Hastie, N. D. Nature 326, 96-99 (1987). 6. Laskowski, M. Jr et al. Biochemistry 26, 202-221 (1987). 7. Brown, A. L. Nature 326, 12-13 (1987). 8. Fleming, A. Lancet 216, 217-220 (1929). 9. Jolles, P. & Jolles, J. Molec. cell Biochem. 63, 165-189 (1984). 10. Bauchop, T. & Martucci, R. W. Science 161, 698-700 (1968). 11. Olivers, D. J. & Hladik, C. M. J. Morph. 166, 337-386 (1980). 12. Padgett, G. A. & Hirsch, J. G. Aust. J. exp. Biol. med. Sci. 45, 569-570 (1967). 13. Pahud, J.-J., Schellenberg, D., Monti, J. C. & Scherz, J. C. Ann. Rech. Vet. 14,493-501 (1983). 14. Prieur, D. J. Comp. Biochem. Physiol. 85B, 349-353 (1986). 15. Wilson, A. C., Carlson, S. S. & White, T. J. A. Rev. Biochem. 46, 573-639 (1977). 16. Goodman, M. Prog. Biophys. molec. Biol. 38, 105-164 (1981). 17. Sarich, V. M. & Cronin, J. E. in Molecular Anthropology (eds Goodman, M. & Tashian, R. E.) 141-170 (Plenum, New York, 1976). 18. Stewart, C.-B., Dobson, D. E. & Wilson, A. C. Am. J. phys. Anthrop. 63, 222 (1984). 19. Haas, O. & Simpson, G. G. Proc. Am. phil. Soc. 90, 319-349 (1946). 20. Zuckerkandl, E. & Pauling, L. in Evolving Genes and Proteins (eds Bryson, V. & Vogel, H. J.) 97-166 (Academic, New York, 1965). 21. Sneath, P. H. A. & Sokal, R. R. Numerical Taxonomy (Freeman, San Francisco, 1973). 22. Peacock, D. & Boulter, D. /. molec. Biol. 95, 513-527 (1975). 23. Weaver, L. H. et al. J. molec. Evol. 21, 97-111 (1985). 24. Creighton, T. E. Proteins: Structures and Molecular Properties (Freeman, New York, 1983). 25. Romero-Herrera, A. E., Lehmann, H., Joysey, K. A. & Friday, A. E. Phil. Trans. R. Soc. B283, 61-163 (1978). 26. Liao, H., McKenzie, T. & Hageman, R. Proc. natn. Acad. Sci. U.S.A. 83, 576-580 (1986). 27. Stewart, C.-B. & Wilson, A. C. Cold Spring Harb. Symp. quant. Biol. 52 (in the press). 28. Rodriguez, R., Menendez-Arias, L., Gonzalez de Buitrago, G. & Gavilanes, J. G. Biochem. Internat. 11, 841-843 (1985). 29. Pervaiz, S. & Brew, K. Arch Biochem. Biophys. 246, 846-854 (1986). 30. Hammer, M. F., Schilling, J. W., Prager, E. M. & Wilson, A. C. / molec. Evol. 24, 272-279 (1987). 31. Felsenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.0 Manual (University of Washington, Seattle, 1987). 32. Jolles, P. et al. J. biol. Chem. 259, 11617-11625 (1984). 33. McKenzie, H. A. & Shaw, D. C. Biochem. Intl 10, 23-31 (1985). 34. Hunkapiller, M. W., Hewick, R. M., Dreyer, W. J. & Hood, L. E. Meth. Enzym. 91,399-413 (1983). 35. Hunkapiller, M. W. & Hood, L. E. Meth. Enzym. 91, 486-493 (1983). 36. Drapeau, G. R. Meth. Enzym. 47, 189-191 (1977). 37. Strydom, D. J. et al. Biochemistry 24, 5486-5494 (1985). 38. Huang, H. V., Bond, M. W., Hunkapiller, M. W. & Hood, L. E. Meth. Enzym. 91, 318-324 (1983). 39. Swofford, D. L. PAUP: Phylogenetic Analysis Using Parsimony, Version 2.4 (Illinois Natural History Survey, Champaign, Illinois, 1985).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Stewart, CB., Schilling, J. & Wilson, A. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 330, 401–404 (1987). https://doi.org/10.1038/330401a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/330401a0
This article is cited by
-
Gaur genome reveals expansion of sperm odorant receptors in domesticated cattle
BMC Genomics (2022)
-
Integration of multi-omics data reveals cis-regulatory variants that are associated with phenotypic differentiation of eastern from western pigs
Genetics Selection Evolution (2022)
-
Identification and expression profile of microRNA in seven tissues of the Golden snub-nosed monkey (Rhinopithecus roxellanae)
Molecular Genetics and Genomics (2020)
-
Duplication and parallel evolution of the pancreatic ribonuclease gene (RNASE1) in folivorous non-colobine primates, the howler monkeys (Alouatta spp.)
Scientific Reports (2019)
-
Convergent genomic signatures of flight loss in birds suggest a switch of main fuel
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.