Network medicine
Network dynamics-based cancer panel stratification for systemic prediction of anticancer drug response
Minsoo Choi et al., Nat. Commun. 8 1940 (2017)
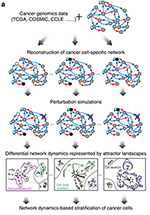 Genomic alterations underlie the variability of drug responses between cancers, but our mechanistic understanding is limited. Here the authors use the p53 network to study how rewiring of signalling networks by genomic alterations impact their dynamic response to pharmacological perturbation.
Genomic alterations underlie the variability of drug responses between cancers, but our mechanistic understanding is limited. Here the authors use the p53 network to study how rewiring of signalling networks by genomic alterations impact their dynamic response to pharmacological perturbation.
Percolation transition of cooperative mutational effects in colorectal tumorigenesis
Dongkwan Shin et al., Nat. Commun. 8 1270 (2017)
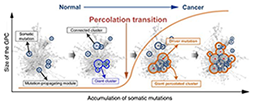 Cancer is caused by accumulating genetic mutations, but their global interactions are poorly understood. Here, the authors investigate the cooperative effect of these mutations in colorectal cancer patients and identify a giant cluster of mutation-propagating modules that undergoes percolation transition from small, scattered modules to a large connected cluster during tumourigenesis.
Cancer is caused by accumulating genetic mutations, but their global interactions are poorly understood. Here, the authors investigate the cooperative effect of these mutations in colorectal cancer patients and identify a giant cluster of mutation-propagating modules that undergoes percolation transition from small, scattered modules to a large connected cluster during tumourigenesis.
Control of primary metabolism by a virulance regulatory network promotes robustness in a plant pathogen
Rémi Peyraud et al., Nat. Commun. 9 418 (2018)
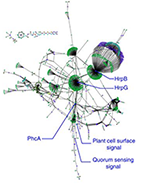 How pathogens maintain phenotypic robustness during infection is poorly understood. Here the authors couple the virulence regulatory network (VRN) of the plant pathogen R. solanacearum to a model of its metabolic network, and find that the VRN activates functionally redundant primary metabolism genes to promote phenotypic robustness during infection.
How pathogens maintain phenotypic robustness during infection is poorly understood. Here the authors couple the virulence regulatory network (VRN) of the plant pathogen R. solanacearum to a model of its metabolic network, and find that the VRN activates functionally redundant primary metabolism genes to promote phenotypic robustness during infection.
A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information
Yunan Luo et al., Nat. Commun. 8 573 (2017)
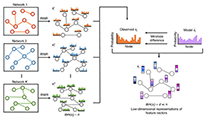 Network-based data integration for drug-target prediction is a promising avenue for drug repositioning, but performance is wanting. Here, the authors introduce DTINet, whose performance is enhanced in the face of noisy, incomplete and high-dimensional biological data by learning low-dimensional vector representations.
Network-based data integration for drug-target prediction is a promising avenue for drug repositioning, but performance is wanting. Here, the authors introduce DTINet, whose performance is enhanced in the face of noisy, incomplete and high-dimensional biological data by learning low-dimensional vector representations.
Network-based in silico drug efficacy screening
Emre Guney et al., Nat. Commun. 7 10331 (2016)
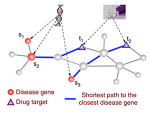 Interaction of a drug with its target protein initiates a perturbation that cascades through the interactome, but this systemic effect is rarely considered in attempts of predicting novel uses of drugs. Here, the authors introduce a measure of drug-disease proximity on the interactome, enabling drug efficacy prediction based on the distance of a drug’s target from the neighborhood in the network that is perturbed in each disease.
Interaction of a drug with its target protein initiates a perturbation that cascades through the interactome, but this systemic effect is rarely considered in attempts of predicting novel uses of drugs. Here, the authors introduce a measure of drug-disease proximity on the interactome, enabling drug efficacy prediction based on the distance of a drug’s target from the neighborhood in the network that is perturbed in each disease.
Centrality in the host–pathogen interactome is associated with pathogen fitness during infection
Núria Crua Asensio et al., Nat Commun. 8, 14092 (2017)
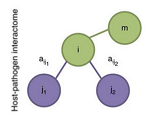 Highly connected nodes in protein-protein interaction networks play essential roles in organismal function. Here, the authors show that it is the proteins with high centrality in the interface of the Y. pestis interacome and that of its host, rather than the pathogen's own protein network, that are most important for pathogen fitness during infection, and highlight the importance of pathogen proteins that likely cause significant perturbation of the interactome of the host.
Highly connected nodes in protein-protein interaction networks play essential roles in organismal function. Here, the authors show that it is the proteins with high centrality in the interface of the Y. pestis interacome and that of its host, rather than the pathogen's own protein network, that are most important for pathogen fitness during infection, and highlight the importance of pathogen proteins that likely cause significant perturbation of the interactome of the host.
Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions
Edik M. Blais et al., Nat. Commun. 8 14250 (2017)
 The rat is a widely used model for human biology, but we must be aware of metabolic differences to judge when it is a good one. Here, the authors reconstruct the genome-scale metabolic network of the rat and reconcile it with an improved human metabolic model. The power of the models to integrate toxicogenomics data is demonstrated by providing species-specific biomarker predictions in response to a panel of drugs.
The rat is a widely used model for human biology, but we must be aware of metabolic differences to judge when it is a good one. Here, the authors reconstruct the genome-scale metabolic network of the rat and reconcile it with an improved human metabolic model. The power of the models to integrate toxicogenomics data is demonstrated by providing species-specific biomarker predictions in response to a panel of drugs.
A geometrical approach to control and controllability of nonlinear dynamical networks
Le-Zhi Wang et al., Nat. Commun. 7 11323 (2016) Complex networks with nonlinear dynamics and multiple stable states, including physical, biological and social systems are ubiquitous, and our ability to control them crucially depends on understanding of how perturbations affects their global dynamics. Here, Wang et al. develop a framework based on the concept of attractor networks to quantify and facilitate the study of controllability of nonlinear dynamics in complex systems. The utility of the framework is demonstrated by finding perturbations that could drive cancerous cell states back to normal states.
Complex networks with nonlinear dynamics and multiple stable states, including physical, biological and social systems are ubiquitous, and our ability to control them crucially depends on understanding of how perturbations affects their global dynamics. Here, Wang et al. develop a framework based on the concept of attractor networks to quantify and facilitate the study of controllability of nonlinear dynamics in complex systems. The utility of the framework is demonstrated by finding perturbations that could drive cancerous cell states back to normal states.
Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury
Jessica L. Nielson et al., Nat. Commun. 6 8581 (2015)
 Precision medicine, particularly for complex neurological disorders, can be enhanced by using data-driven discovery to extract meaningful knowledge from large, heterogeneous datasets. Here, the authors apply a big-data analytical approach known as Topological Data Analysis (TDA) to assess therapeutic effects in preclinical traumatic brain injury and spinal cord injury research studies. With this approach, they identify novel symptomatic patterns, detrimental drug effects, and predict long-term recovery.
Precision medicine, particularly for complex neurological disorders, can be enhanced by using data-driven discovery to extract meaningful knowledge from large, heterogeneous datasets. Here, the authors apply a big-data analytical approach known as Topological Data Analysis (TDA) to assess therapeutic effects in preclinical traumatic brain injury and spinal cord injury research studies. With this approach, they identify novel symptomatic patterns, detrimental drug effects, and predict long-term recovery.