Abstract
Chronic lymphoid leukemia (CLL) is characterized by the accumulation of functionally defective CD5-positive B lymphocytes. The clinical course of CLL is highly variable, ranging from a long-lasting indolent disease to an unpredictable and rapidly progressing leukemia requiring treatment. It is thus important to identify novel factors that reflect disease progression or contribute to its assessment. Here, we report on a novel STAT3-mediated pathway that characterizes CLL B cells-extended viability and oxidative stress control. We observed that leukemic but not normal B cells from CLL patients exhibit constitutive activation of an atypical form of the STAT3 signaling factor, phosphorylated on serine 727 (Ser727) in the absence of detectable canonical tyrosine 705 (Tyr705)-dependent activation in vivo. The Ser727-phosphorylated STAT3 molecule (pSTAT3Ser727) is localized to the mitochondria and associates with complex I of the respiratory chain. This pSer727 modification is further controlled by glutathione-dependent antioxidant pathway(s) that mediate stromal protection of the leukemic B cells and regulate their viability. Importantly, pSTAT3Ser727, but neither Tyr705-phosphorylated STAT3 nor total STAT3, levels correlate with prolonged in vivo CLL B cells survival. Furthermore, STAT3 activity contributes to the resistance to apoptosis of CLL, but not normal B cells, in vitro. These data reveal that mitochondrial (Mt) pSTAT3Ser727 overactivity is part of the antioxidant defense pathway of CLL B cells that regulates their viability. Mt pSTAT3Ser727 appears to be a newly identified cell-protective signal involved in CLL cells survival. Targeting pSTAT3Ser727 could be a promising new therapeutic approach.
Similar content being viewed by others
Main
Chronic lymphoid leukemia (CLL) is characterized by the accumulation of proliferating CD5+ B lymphocytes.1 The clinical course of CLL is highly variable, ranging from a long-lasting to a rapidly progressing leukemia requiring treatment. Although the understanding of CLL pathophysiology has improved markedly in the past few years, therapeutic approaches are still not curative. It is thus important to further identify the factors that selectively sustain CLL cells survival.
CLL B lymphoid cells (CLL-BCs) have a prolonged survival time in vivo but exhibit a high level of spontaneous apoptosis in vitro, highlighting the critical role of CLL-BC microenvironment in this pathology.1-3 Several stromal factors have been shown to be implicated in promoting CLL-BC viability in vitro,4, 5, 6 yet the exact mechanisms by which the microenvironment protects CLL cells from apoptosis in vivo remain unclear.
The STAT3 signaling factor mediates numerous extracellular survival/growth messages. It is activated by phosphorylation of tyrosine 705 (Tyr705), allowing STAT3 to bind to DNA and activate the transcription of target genes.7 Abnormal constitutive activation of Tyr705-phosphorylated STAT3 (pSTAT3Tyr705) is observed in multiple tumor cells and contributes to oncogenic processes.8,9 In addition, STAT3 can be phosphorylated at serine 727 (Ser727) by growth factor-activated serine kinases, thereby modulating STAT3 transcriptional activity and regulating the activity of associated transcriptional factors such as NFκB.10 Remarkably, STAT3 was recently reported to exhibit extranuclear pro-oncogenic activities in murine cells, linked to its mono-phosphorylation on Ser727 but not Tyr705, subsequent association with mitochondrial (Mt) components and regulation of the respiratory chain.11,12 In 1997, Frank and Mahajan13 showed by western blotting a remarkable constitutive phosphorylation of STAT3Ser727 in the absence of canonical pSTAT3Tyr705 in 100% of 32 primary CLL-BC samples. Hazan-Halevy et al.14 later showed that STAT3 limited the spontaneous apoptosis of CLL-BC in vitro.
In the present study, we addressed the role of Ser727-phosphorylated STAT3 (pSTAT3Ser727) in CLL pathophysiology. We demonstrate that circulating CLL-BCs but not normal B lymphoid cells (N-BCs) overexpress an atypical mitochondria-associated pSTAT3Ser727 moiety, the level of which correlates with prolonged CLL-BC survival in vivo. We further identified glutathione (GSH), an antioxidant mediating stromal protection of the leukemic cells, to be a critical regulator of Ser727 phosphorylation, and provide evidence that STAT3 is responsible for the extended survival of CLL-BC as compared with N-BC. Overall, our data suggest that Mt pSTAT3Ser727 contributes to the protection of CLL-BC. As such, it may represent a new biochemical pathway for effectively targeting leukemia cells, especially those that exhibit drug resistance.
Results
Overactivation of pSTAT3Ser727 correlates with CLL-BC resistance to apoptosis
Circulating B cells were purified from healthy donors (n=8) and CLL patients (Supplementary Table 1). Samples were studied immediately upon collection to avoid any culture artifacts owing to the spontaneous apoptosis of these cells in vitro. Apoptosis was measured by combining Annexin V/7-AAD (7-aminoactinomycin D) staining of phosphatidyl serine externalization and dead cell DNA labeling. Apoptosis was also assessed by monitoring the Mt transmembrane potential (MTP) that characterizes mitochondria in viable cells. CLL-BC showed significantly lower apoptosis than N-BC using both assays, consistent with the extended survival of CLL-BC in vivo (Figure 1a and data not shown). The intensity of this labeling was variable from patient to patient, which is in agreement with the heterogeneity of CLL disease. STAT3 phosphorylation was investigated by flow cytometry measurements (FCMs). As shown in Figure 1b, CLL-BC showed strong immunolabeling using an anti-pSTAT3Ser727 antibody. This labeling was abolished by the pSTAT3Ser727 immunogen peptide (ipep) as well as by an 11 amino-acid-long STAT3Ser727 phosphopeptide but not by the unphosphorylated peptide, thus confirming labeling specificity. Conversely, quite low pSTAT3Ser727 immunolabeling was detected in N-BC under similar conditions, although pSTAT3 Ser727 was normally induced by phorbol esters in these cells (Figure 1b, left panel). Statistical analyses confirmed that circulating CLL-BC expressed higher levels of pSTAT3Ser727 as compared with N-BC (Figure 1c).
Activation of pSTAT3Ser727 correlates with CLL-BC resistance to apoptosis. (a) Flow cytometry measurement (FCM) of B-cell apoptosis by Annexin V staining of CD45+CD19+ normal (N-BC) and CLL (CLL-BC) B cells. (b–e) FCM of pSTAT3Ser727 (b and c), STAT3Tyr705 (d) and total STAT3 (e) of N-BC and CLL-BC. (b) One representative experiment is shown. B cells were labeled with rabbit control (dashed black line), total STAT3 or pSTAT3Ser727 antibodies, as indicated, in the absence (blue line) or presence of pSTAT3Ser72 immunogen peptide (ipep; green line), phosphorylated S3Ser727 11-peptide (red line) or unphosphorylated S3Ser727 11-peptide (purple line). The black line indicates pSTAT3Ser727 labeling following a 15-min treatment of N-BC with phorbol myristyl acetate. Results are expressed as mean fluorescence intensity (arbitrary unit, a.u.). (c–e). Statistical analyses of the indicated immunolabeling mean fluorescence intensities are shown (N-BC, n=8; CLL-BC, n=29; **P<0.01). (f and g) Correlation between pSTAT3Ser727 or total STAT3 levels (mean fluorescence intensity) and the percentage of apoptotic Annexin V-positive cells (f) or DiOC6-probed mitochondrial transmembrane potential (G), evaluated in CLL-BC samples (r, Pearson’s coefficient; n=29)
Regarding pSTAT3Tyr705, CLL-BC and N-BC showed a similar weak-to-undetectable immunolabeling (Figure 1d). In addition, CLL-BC and N-BC expressed the same total STAT3 levels (Figure 1e). Western blot analysis of the same cells confirmed that CLL-BC overexpressed pSTAT3Ser727 as compared with N-BC in the absence of detectable pSTAT3Tyr705. Also, no activation of the related STAT5 factor was observed (Supplementary Figure 1).
The highly variable course of CLL led us to compare pSTAT3Ser727 levels with the apoptosis indexes of CLL-BC. A significant negative correlation was observed between pSTAT3Ser727 level and the percentage of apoptotic Annexin V-positive CLL-BC (Figure 1f, right, n=29). Conversely, pSTAT3Ser727 levels showed a positive correlation with the mitochondria transmembrane potential (MTP) that labeled viable cells (Figure 1g, right, n=29). Notably, no correlation was observed between total STAT3 levels and CLL-BC basal apoptosis using both criteria. These data indicate that CLL-BC-extended survival correlates with pSTAT3Ser727 activation in the absence of pSTAT3Tyr705 activation and total STAT3 amplification. This suggests that pSTAT3Ser727 contributes to CLL-BC leukemic properties in vivo.
pSTAT3Ser727 localizes to CLL-BC mitochondria
A single serine phosphorylation was shown to mediate STAT transcription activation.15 We determined whether CLL-BC pSTAT3Ser727 binds DNA. CLL-BC nuclear extracts were prepared and pull-down experiments using biotin-labeled STAT3-DNA probes were performed. pSTAT3Ser727 was mostly undetectable in the nuclear fraction. Neither STAT3 nor pSTATSer7227 did bind STAT3-DNA elements (Figure 2a and data not shown). pSTAT3Ser727 localization was next examined using immunolabeling coupled to confocal microscopy. In CLL-BC, pSTAT3Ser727 distributed to the cytoplasm with a clear granular pattern but no significant nuclear labeling (Figure 2b, top). All fluorescence was suppressed when the rabbit anti-pSTAT3Ser727 antibody was pre-mixed with an excess amount of ipep (Figure 2b) or with pS3-11pep but not with the unphosphorylated STAT3 (S3)-11pep (Supplementary Figure 2). Compared with CLL-BC, N-BC showed weak-to-undetectable pSTAT3Ser727 labeling (Figure 2b). Immunolabeling of total STAT3 showed a rather uniform cellular distribution in CLL-BC and N-BC (Figure 2b). The peculiar intracellular distribution of pSTAT3-Ser727 suggests an association with cytoplasmic organelles. Because of the reported Mt activities of STAT3,12 the important involvement of mitochondria in cell survival and the increased MTP of CLL-BC (Figure 1), we focused on these organelles. Mitotracker, a selective dye that concentrates into mitochondria along MTP, was first used to visualize these organelles. pSTAT3-Ser727 was observed within Mitotracker-positive subcellular areas in co-labeling assays (Figure 2b). An additional experiment was performed using a murine monoclonal anti-pSTAT3Ser727 antibody that confirmed the predominant granular cytoplasmic localization of pSTAT3Ser727 in CLL-BC (Figure 2c). Mitochondria were co-labeled by antibodies directed to the Mt electron transfer chain (ETC) complex I NADH-ubiquinone reductase subunit (mtND1).16 pSTAT3-Ser727 immunolabeling again co-localized with this second Mt marker, as assessed by line-scan fluorescence quantification (Figure 2c). Co-labeling was further ascertained at the pixel level using Pearson’s correlation coefficient measurement that validated pSTAT3Ser727/mitochondria co-localization statistically in patient samples (Figure 2d).
pSTAT3Ser727 subcellular localization characterizes human primary CLL B cells. (a) Nuclear extracts from CLL-BC (#n upper panel, lower panel) or UT7 hematopoietic cell line were prepared and incubated with biotin-labeled STAT3-binding site DNA probes and streptavidin magnetic beads for pull-down assays. Attached proteins were separated by SDS-PAGE and STAT3 was detected by immunoblotting. NE and OP indicate nuclear extracts and oligo-pull-down extracts, respectively; 1:100 of total NE were loaded on each gel. NE were incubated with biotin-labeled SIEm67- (upper panel) or with SIEm67 (S), IRF (I)- or Oct (O)-DNA (lower panel) probes; lo and hi indicate low and high exposure of the immunoblot. UT7 hematopoietic cells were pre-incubated for 15 min in the absence or presence of phorbol myristyl acetate (P) as positive controls. α and β indicate STAT3α and STAT3β isoforms. (b–d) CLL-BC and N-BC were isolated, fixed, permeabilized and processed for fluorescence confocal microscopy as indicated. (b) Rabbit (rb) anti-pSTAT3Ser727 (pS3Ser727) antibody in the absence or presence of immunogen peptide (ipep), rabbit anti-total STAT3 (total S3) and isotype control rabbit immunoglobulins (rb Ig) were used in co-labelings with mitotracker, as indicated. DIC indicates differential interference contrast. (c) Mouse anti-pSTAT3Ser727 (pS3Ser727 mAb) and rabbit anti-mtND1 antibodies were used in co-labeling assays. Fluorescent probes were FITC-coupled anti-rabbit Ig (rb Ig-FITC, green), red-mitotracker or PE-coupled anti-mouse Ig (red), DAPI (blue). Line scan intensity profiles (bottom) of pS3Ser (red), mtND1 (green) and DAPI (blue) labeling along indicated axes (x1–y1; x2–y2) from gated areas. DAPI-positive nuclear (N) and DAPI-negative cytoplasmic (C) areas are schematized. Representative images from five patients (#n relates to Table I patient’s number) and five healthy donor samples are shown. Magnification, × 100. (d) Pearson’correlation coefficients were measured for the indicated fluorescent signals (rabbit anti-pSTAT3Ser727 versus mitotracker; mouse anti-pSTAT3Ser727 versus rabbit anti-mtND1; mouse anti-pSTAT3Ser727 versus DAPI). Twenty-five cells were randomly examined in confocal microscopy for each patient, and data from five and three patients were pooled, which corresponds to over 100 cells observed; **P<0.01)
B lymphocytes are small cells with a very limited cytoplasmic area. pSTAT3Ser727 intracellular distribution was further analyzed using transmission electron microscopy. As shown in Figure 3a, CLL-BC contained significantly more mitochondria as compared with N-BC (10.6±0.45 versus 7.5±0.17, P<0.01), which filled most of the cytoplasmic area. Immunogold labeling, performed on ultrathin sections, revealed the presence of immunoreactive pSTAT3Ser727, predominantly localized in CLL-BC mitochondria (Figure 3b). No signal was detected in N-BC nor in CLL-BC stained in the presence of the ipep (data not shown). By contrast, immunolabeling of total STAT3 revealed the presence of STAT3 moieties in all cell compartments, including mitochondria, in both CLL-BC and N-BC (Figure 3c).
pSTATSer727 associates with mitochondria of CLL-BC but not N-BC. (a) CLL-BC (n=10) and N-BC (n=2) were processed for transmission electron microscopy (TEM) as described. Left panel: representative images of B-cell ultrastructures. Right panel: quantification of mitochondria numbers per TEM cell section. Data are presented as mean±S.E.M., (**P<0.01). (b and c) CLL-BC (n=3) and N-BC (C, n=2) were fixed, permeabilized and processed for immunogold labeling and TEM using rabbit pSTAT3Ser727 (b) or total STAT3 (c) antibody, as indicated. White and black arrow heads, respectively, indicate cytosol and plasma membrane (PM)- localized signals; dashed black arrows indicate nuclear (N) signals; white arrows indicate signals associated to mitochondria
pSTAT3Ser727 intracellular localization was also assessed by subcellular fractionation. Mitochondria were isolated from cell lysates by magnetic labeling with antibodies directed to the translocase of the outer Mt membrane 22 (TOM22). Mitochondria enrichment was evaluated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) coupled to immunoblotting using the Mt heat-shock protein 60 (hsp60) and NADH dehydrogenase 1 alpha subunit 9 (NDUFA9), a component of complex I of the ETC, cytoplasmic β-actin and nuclear lysine demethylase 1a (KDM1a; Figure 4a). STAT3 and pSTAT3Ser727 associate with β-actin- and KDM1a-negative mitochondria-enriched fraction. Comparison of the amounts of STAT3 and hsp60 in TOM22-positive and -negative fractions indicated that levels of Mt STAT3 were ~1–5% that of total STAT3. First, evidence for the Mt localization of STAT3 has come from observations that GRIM-19, a component of the ETC complex I, directly binds to STAT3.12 Using an antibody that captures all components of complex I and immunoprecipation assays, we co-precipitated serine phosphorylated STAT3 in CLL-BC, but not in N-BC extracts (Figure 4b).
pSTATSer727 associates with complex I of the mitochondria respiratory chain. (a) Mitochondria (Mt) were isolated from CLL-BC (whole cells, WC) by magnetic immunolabeling. Extracts were separated by SDS-PAGE and blots were probed with the indicated antibodies. (b) Antibody to complex I (CI) of the ETC or a nonspecific isotype-matched IgG (ctl) were incubated with CLL-BC or N-BC total cell extracts (TEs), as indicated. Immunoprecipitates (IPs) were resolved on SDS-PAGE and probed for the indicated proteins. 1 : 50 of TE was loaded on the gel. α and β indicate STAT3α and STATβ isoforms, respectively
Collectively, these data concur in indicating that CLL-BC display overactivation of an atypical Mt pSTAT3Ser727 moiety.
CLL-BC oxidative stress regulates Mt pSTAT3Ser727 activation
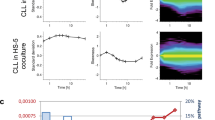
In vivo, their microenvironment has a profound effect on CLL-BC survival. Stromal support of CLL-BC can be reproduced when they are co-cultured with bone marrow stromal cell lines.6,17 Indeed, murine MS5 and human HS5 cells significantly reduce the level of CLL-BC spontaneous apoptosis, as assessed by Annexin V labeling and MTP probing (Figures 5a and b and Supplementary Figure 3). It similarly maintains a strong level of pSTAT3Ser727 immunolabeling in absence of any immunoreactive pSTAT3Tyr705 in CLL-BC even after prolonged periods (Figures 5c and d, right panels, and Supplementary Figure 3), while the expression of total STAT3 remains unchanged (data not shown). Under similar conditions, stromal cells enhance not only pSTAT3Ser727 but also pSTAT3Tyr705 activation of N-BC in a time-dependent manner.
Stromal protection of CLL-BC sustains Mt pSTAT3Ser727 overactivation. (a and b) FCM of apoptosis (a) and mitochondrial transmembrane potential (MTP; b) of N-BC and CLL-BC at day 8 of MS5 co-culture. Results are expressed as percentage of AnnexinV-positive (a) and TMRM-positive (MTP+; b) cells. The mean +/− S.E.M. of 3 independent experiments is shown, *P<0.05; **P<0.01. (c and d) FCM analysis of pSTAT3Ser727 (c) or pSTAT3Tyr705 (d) expression in N-BC and CLL-BC upon MS5 co-culture. CD45+CD19+ CD5+ B cells were analyzed at day 0 (blue), day 3 (purple) or day 8 (red) of MS5 co-culture. Standard rabbit immunoglobulins were used as control at day 8 (dashed black line). One representative experiment is shown (samples from three healthy donors and three individual patients were analyzed). Results are expressed as mean fluorescence intensity (arbitrary unit, a.u.). (e) N-BC and CLL-BC were collected at day 8 of MS5 co-culture. Immunofluorescence analyses were performed with the indicated primary antibodies using fluorescence microscopy (magnification × 100). Fluorescent probes were FITC-coupled anti-rabbit Ig (green), PE-coupled anti-mouse Ig (red) and DAPI (blue). DIC indicates differential interference contrast. Representative images from three CLL patients and five healthy donors are shown
pSTAT3Ser727 localization was assessed in this model. Using again two different pSTAT3Ser727 antibodies and mitochondria markers, CLL-BC were shown to display the same predominant Mt localization of pSTAT3Ser727 upon MS5 co-culture, as compared with freshly isolated CLL-BC (Figure 5e and data not shown). Conversely, a pronounced difference was observed in stroma co-cultured N-BC in which pSTAT3Ser727 was distributed to the cytoplasmic and nuclear compartments, and thus not predominantly associated to mitochondria.
Collectively, these observations indicate that stromal cells mimic in vitro the interactions between CLL-BC and their microenvironment that occur in vivo, in terms of CLL-BC protection from apoptosis, STAT3 phosphorylation status and intracellular localization. They further reveal that the same stromal messages are differentially received and translated by N-BC and CLL-BC, thereby leading to mitochondria-restricted pSTAT3Ser727 overexpression in leukemic cells only.
The accumulation of Mt pSTAT3Ser727 in stroma-supported CLL-BC led us to search for stromal signals that would regulate pSTAT3Ser727 activation in these cells. CLL-BC intrinsically have high levels of reactive oxygen species (ROS) when compared with normal lymphocytes.1,18 This makes CLL-BC more dependent on such cellular antioxidants as GSH. A critical metabolic interaction between CLL-BC and bone marrow stromal cells was recently reported to enhance GSH synthesis, and thus increase the ability of CLL-BC to maintain the redox balance and promote cell survival.6 We tested the relationship between GSH, oxidative stress and pSTAT3Ser727 expression of CLL-BC. Dihydroethidium (DHE) provides a clear indication of primary ROS levels19 by binding to superoxide anions that are produced predominantly by the respiratory chain of mitochondria in most cells except phagocytes. As shown in Figure 6a, the addition of GSH to CLL-BC culture medium, in absence of stromal cells, indeed enhanced ROS detoxification, thereby lowering superoxide anions levels and promoting cell survival. It further enhanced pSTAT3Ser727 expression of CLL-BC without changing pSTAT3Tyr705 activation (data not shown) and total STAT3 levels. Similar data were obtained upon addition of N-acetylcysteine (NAC), another antioxidant. This increased activation of pSTAT3Ser727 by antioxidants targeted Mt STAT3, as assessed by immunolabeling coupled to confocal microscopy (data not shown).
GSH metabolism regulates the phosphorylation of STAT-Ser727 of CLL-BC. (a and b) Comparison of apoptosis, ROS, pSTAT3Ser727 and total STAT3 expression of CLL-BC cultured alone for 4 days in the presence or absence of NAC (1 mM) or GSH (2 mM). Where indicated, PEITC (1–5 μM, 5 h) was added at the end of the culture. (c) Time course of ROS, pSTAT3Ser727 and total STAT3 expression of CLL-BC cultured alone in the presence of PEITC (5 μM) for the indicated time. ROS were measured by DHE staining of CD45/CD19+/CD5+ cells; pSTAT3Ser727 and total STAT3 levels were evaluated upon immunolabeling/FCM. Data are expressed as relative to untreated cells. The mean±S.E.M. of four separate experiments with four different CLL patient samples are shown (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001). (d) FCM of pMEK (right) and pSTAT3Ser727 (left) of CLL-BC upon a two-day culture in the absence (black) or presence of 10 nM (dark grey) or 100 nM (medium grey) of MEK inhibitor PD0325901. B cells were labeled with rabbit control (light grey), pMEK or pSTAT3Ser727 antibodies, as indicated. Results are expressed as mean fluorescence intensity (arbitrary unit, a.u.)
Conversely, the viability of CLL cells can be decreased by depletion of GSH using β-phenylethyl isothiocyanate (PEITC), a natural compound capable of inactivating cellular GSH and modulating ETC activity. PEITC indeed significantly enhanced intracellular ROS levels of CLL-BC (Figure 6b). It also decreased CLL-BC levels of pSTAT3Ser727 in a concentration-dependent manner while it had no impact on total STAT3 expression. It further suppressed GSH- and NAC-dependent regulation of ROS and pSTAT3Ser—but again not total STAT3–expression. Notably, the inhibitory activity of PEITC was time dependent, with a significant pSTAT3Ser727 loss being detectable at 1 h of treatment. This effect was observed when CLL-BCs were cultured alone (Figure 6c) or with stromal cells (data not shown), and were detectable before substantial ROS increase. These data suggest that the oxidative stress status of CLL-BC regulates pSTAT3Ser727. ROS directly affects the activity of a large number of cellular proteins. It also regulates the amounts of reduced GSH, which in turn positively or negatively modulate protein activity through direct conjugation, that is, glutathionylation, as observed for such Ser/Thr kinases as MEK and JNK, which may mediate GSH-dependent Mt STAT3Ser727 phosphorylation.20 The MEK–ERK pathway is required for Ras-induced phosphorylation of Mt STAT3 on Ser727, which is one of the critical steps of the Ras-MEK–ERK axis during transformation.21 Pharmacological inhibition of MEK, however, did not affect serine phosphorylation of STAT3 in CLL-BC (Figure 6d).
Altogether, these data suggest that the oxidative stress status of CLL-BC regulates pSTAT3Ser727 activation through a MEK–ERK independent pathway.
STAT3 sustains CLL-BC survival
The role of STAT3 in mediating the protection of CLL-BC was next investigated by an RNA interference-based strategy. Small hairpin RNA (shRNA) that knocks down human STAT3 (shS3) was stably introduced in N-BC and CLL-BC, using a lentiviral vector that co-encodes the green fluorescent protein (GFP) dye. Lentiviral particles that expressed measles virus glycoprotein envelope instead of canonical vesicular stomatitis virus glycoprotein were used to allow efficient transduction of human B cells, as we reported previously.22 shRNA targeting luciferase (shL) or STAT5 (shS5) were used as controls. As shown in Figures 7a and b, shS3 decreased STAT3 gene expression, with no impact on STAT5 levels as compared with control shRNA (shL)-expressing human RAJI B cells, which validated the shS3 specificity. This was confirmed on CLL-BC by FCM. Transduced CLL-BC were maintained on MS5 stromal cells and the level of apoptosis of GFP+ and GFP− subsets were analyzed along the period of culture, using AnnexinV labeling and tetramethylrhodamine-methyl-ester (TMRM) MTP probing, together with dead cell DNA staining by 7AAD or TOPRO-3 dyes. By all criteria, shS3+/GFP+ CLL-BC exhibited enhanced apoptosis as compared with shS3-/GFP- and shL+/GFP+ control cells, whereas the viability of shS5+/GFP+ CLL-BC was unchanged (Figures 7c–e). Notably, shS3 suppressed the enhanced viability of CLL-BC, whereas it had no consequences on N-BC under similar conditions (Figures 7d and e). These data indicate that STAT3, but not STAT5, selectively supports CLL-BC survival in the absence of canonical pSTAT3Tyr705 activation.
CLL-BC depends on STAT3 activity for their growth. (a) Immunoblot analysis of the indicated proteins in shRNA/GFP+ Raji B cells at day 8 post transduction. shL, shS3 and shS5 refer to control luciferase, STAT3 and STAT5 shRNA, respectively. Numbers at the bottom of each panel indicate STAT3/ β-actin and STAT5/β-actin ratio relative to shL cells, respectively. (b) FCM of pSTAT3Ser727 expression in CLL-BC at day 8 post transduction with the indicated shRNA/GFP encoding lentiviral vectors. B cells were co-cultured with MS5 stromal cells since day 1 post transduction. Vertical bars delineate specific fluorescent signals compared with control isotype immunoglobulins (ctl Ig, left panel). Results are expressed as mean fluorescence intensity (arbitrary unit, a.u.). Percentages of pSTAT3Ser727/GFP+ cells are indicated inside each graph. (c–e) N-BC and CLL B cells were transduced with the indicated shRNA/GFP encoding lentiviral vectors. Apoptosis was analyzed at day 8 post transduction, on GFP+ and GFP- B cells by FCM using Annexin V/7-AAD and TMRM/TOPRO3 labeling, as indicated. (c) Representative experiment (n=10), numbers indicate the percentages of cells in relevant quadrants. (d and e) Statistical analyses expressed as percentages of AnnexinV positive (d) and TMRM-probed MTP positive (MTP+) (e) cells. UT indicates untransduced cells (mean±S.E.M., CLL-BC, n=10, N-BC, n=5; **P<0.01; *P<0.05)
Discussion
In the present study, we investigated the physiopathological role of STAT3 phosphorylation in the natural history of CLL disease. Using human primary B cells from healthy donors and CLL-BCs from patients, this study shows that CLL-BC but not N-BC exhibit an over-activation of an atypical mitochondria-associated pSTAT3Ser727 moiety that characterizes leukemic cells survival and oxidative stress control. Four key results support these conclusions. First, circulating CLL-BC spontaneously express an abnormally high level of the mitochondria-restricted pSTAT3Ser727 moiety in the absence of canonical pSTAT3Tyr705 activation, at variance with what is observed in N-BC in vivo. Second, stroma-supported CLL-BCs, but not N-BCs, exhibit overactivation of the Mt pSTAT3Ser727 moiety in vitro. Third, both inhibitors and activators of the antioxidant GSH pathway, which regulates the redox balance and survival of CLL-BC, similarly regulate CLL-BC Mt pSTAT3Ser727. Fourth, the extended survival of CLL-BC correlates with their pSTAT3Ser727 status but not overall STAT3 expression level in vivo and rely on STAT3 activity, as opposed to what was observed in N-BC.
Several reports have shown that pSTAT3Ser727 activation in the absence of canonical pSTAT3Tyr705 mediates cell survival messages, as illustrated by the action of neutrotrophins on both neuronal stem/progenitor and mature cells, or VEGF activity on CLL-BC cultured alone.23, 24, 25, 26 DNA damage inducers similarly activate pSTAT3Ser727 but not pSTAT3Tyr705 expression in cancer cell models, thereby enhancing DNA repair.27 In these analyses, pSTAT3Ser727 activity involved nuclear transcriptional regulation of cell survival/DNA repair genes. Our study reveals that CLL-BC exhibit a unique Mt pSTAT3Ser727 overactivation not observed in N-BC. Wegrzyn et al.12 have indicated that Mt pSTAT3Ser727 enhanced mitochondria respiration of murine normal primary B cells, yet without impacting their survival. It however protected murine cardiomyocytes from ischemia and decreased the production of ROS and cytochrome-c release.28 Also, Mt pSTAT3Ser727 is involved in nerve growth factor-induced neurite outgrowth and the production of ROS.29 Remarkably, Mt pSTAT3Ser727 is overexpressed by Ras-transformed mouse embryo fibroblasts and supports Ras-dependent malignant transformation.11 It further influences the tumorigenic potential of murine 4T1 breast cancer cells that correlates with the regulation of ROS concentrations.30
Here our observations consistently disclosed that human primary CLL-BC spontaneously exhibit overactivation of Mt pSTAT3Ser727 as compared with their normal counterparts, a feature that is linked to their extended survival and oxidative stress control. To our knowledge CLL is the first human malignancy linked to a non-canonical mitochondria-restricted pSTAT3Ser727 overactivity in vivo. One prominent biochemical feature of CLL-BC is their high level of ROS and oxidative stress, when compared with normal lymphocytes. This ROS stress renders them dependent on proper redox balance and makes them oversensitive to agents that induce further ROS stress.31,32 Interestingly, we observed that CLL-BC exhibit an abnormal accumulation of Mt mass, which is in agreement with the data reported by Carew et al.18 The deregulated mitochondria biogenesis in CLL-BC is significantly related to endogenous ROS levels and further correlates with enhanced drug resistance.18 GSH is the most abundant antioxidant in cells and significantly affect cell survival.33 Indeed, CLL-BC from patients exhibit deficient GSH synthesis and rely on stromal cells to provide cysteine for GSH production.6 Our study reveals that the oxidative stress status of CLL-BC, which is controlled by cell microenvironment and dictates leukemic cell apoptosis resistance, also regulates the activation of Mt pSTAT3Ser727 in CLL-BC in a dose-dependent and rapid manner (<1 h), and also in absence of the active MEK–ERK pathway. It indicates that various signaling pathways can modulate serine phosphorylation of Mt STAT3 in tumor cells.
We have tested a number of inhibitory agents to decipher which kinase(s) might phosphorylate STAT3Ser727 of CLL-BC. They included PI3k inhibitors (Ly294002, Wortmanin), mTOR inhibitors (rapamycin and AZD8055) and many others (data not shown). GSH activators and inhibitors were the very first ones to affect pSTAT3Ser727 of CLL-BC. Clearly, more work needs to be done for pSTAT3Ser727 mode of activation in CLL-BC. Our observation that the GSH antioxidant pathway regulates pSTAT3Ser727 expression questions whether pSTAT3Ser727 effectors may influence ROS production. pSTAT3Ser727 associates with complex I of CLL cells. In the presence of increased ROS, STAT3 can be oxidized to form multimers.34 An alternative oxidative modification of STAT3 by S-glutathionylation has also been observed.35 Complex I-associated STAT3 of CLL-BC may thus limit ROS production by decreasing electron leak of complex I and/or by serving as a critical electron scavenger, as previously suggested.28, 29, 30
In vivo, the microenvironment has a profound effect on CLL-BC survival. Besides BCR signaling, several stromal factors have been implicated in sustaining CLL-BC survival. A number of these factors activate not only pSTAT3Tyr705 but also pSTAT3Ser727 in N-BC and CLL-BC when added to culture medium separately in the absence of stromal cells.4,5,23 Our study now reveals that the mixed protective signals provided by stromal microenvironments in vivo and ex vivo (i.e., MS5/HS5) are all combined to a unique mitochondria-associated overactivation of pSTAT3Ser727 in the absence of canonical activation of pSTAT3Tyr705 in CLL-BC but not N-BC. It provides substantial evidence that the same stromal signals are differentially translated by CLL-BC and N-BC. Thus, stromal support of leukemia cells correlates with major intrinsic qualitative changes in CLL-BC signaling pathways leading to pSTAT3Ser727 overactivation, as compared with N-BC. Our data show that this pSTAT3Ser727 overactivation is not associated with massive STAT3 delocalization. Indeed, immunolabelings of human normal and leukemic B cells detected the presence of reasonable levels of Mt STAT3, which is confirmed by subcellular fractionation. This suggests that constitutive phosphorylation of STAT3Ser727 occurs within the mitochondria of CLL-BC. As opposed to our data, previous reports showed that a minor proportion of total STAT3 localizes to mitochondria upon fractionation of normal cells.11,12,36 One interpretation of this discrepancy is that part of Mt STAT3 is lost upon mitochondria isolation based on different subcellular fractionation protocols. Also, CLL-BC contain significantly more mitochondria as compared with N-BC. Mt STAT3 of CLL-BC might therefore be more abundant and/or less labile as compared with other cells. Also, our data indicate that nuclear pSTAT3Ser is undetectable and unable to bind STAT3-DNA-specific probes in CLL-BC. This is in contradiction with the previously reported nucleocytoplasmic trafficking of pSTAT3Ser727 in CLL-BC, despite the use of an identical STAT3-DNA probe.14 Again, different subcellular fractionation and/or pull-down protocols may explain such a discrepancy.
Overall, our findings indicate that Mt pSTAT3Ser727 overactivation is linked to leukemic B cell-extended survival and antioxidant defense. It suggests that pSTAT3Ser727 contributes to the antioxidant defense and protection of the leukemic cells. This activity may similarly reduce that of ROS-generating anticancer agents and favor drug resistance. These results suggest that pSTAT3Ser727 detection could be used as a new tool to characterize patients’ leukemic cells from blood samples, which, together with current disease criteria, could help define new patient subgroups and help predict disease outcome and/or drug resistance. It further supports the idea of therapeutically targeting pSTAT3Ser727 activity to deregulate leukemic cell antioxidant defense and eliminate the CLL-BC protected by their microenvironment, which could be resistant to immune chemotherapy. To our knowledge, CLL is the first human malignancy linked to a non-canonical mitochondria-restricted pSTAT3Ser727 overactivity in vivo, thereby offering a new potential therapeutic target that spares healthy B cells.
Materials and Methods
Primary cells and cell lines
After obtaining the approved informed consent of Institutional Review Board, peripheral blood mononuclear cells were collected from healthy donors and CLL patients, and were isolated by density-gradient centrifugation on Ficoll Hypaque. B cells were further purified using CD3/CD14 immunomagnetic-bead-negative selection (Miltenyi Biotec, Cologne, Germany) to avoid B-cell activation. CD5/CD19 double-positive cells were selected by FACS. All patients had immunophenotypically defined CLL as outlined by the modified 1996 National Cancer Institute Criteria (Supplementary Table 1). The RAJI human B-cell line, the MS5 murine and HS5 human bone marrow stromal cell lines were purchased from DSMZ cell depository bank. They were cultured in RPMI (RAJI) and MEM alpha (MS5, HS5) medium supplemented with 10% fetal calf serum (FCS). B cells were cultured in vitro alone or with a layer of HS5 or MS5 cells under normoxia in RPMI medium supplemented with 5–10% FCS. The cells were incubated with GSH (2 mM) or NAC (1 mM) for 4 days and exposed to PEITC (0–5 μM) during the last 5 h of incubation. Alternatively, B cells were exposed to PEITC (0–5 μM) alone for 0–5 h. Where indicated, CLL-BC were cultured alone for 2 days in the absence or presence of the MEK inhibitor PD0325901 (0–100 nM).
Lentiviral constructs and transduction
Oligonucleotides targeting firefly luciferase (shL, 5′-CGTACGCGGAATACTTCGA-3′), STAT3 (shS3, 5′-AAGAAACTGGAGGAGTTGCAG-3′) and STAT5A/B (shS5, 5′-GGAGAACCTCGTGTTCCTG-3′) were inserted downstream of the H1 promoter into an HIV-SIN-SFFVGFP vector that encodes GFP as reporter. Lentiviral particles displaying measles virus glycoproteins were produced and titrated on 293T cells using GFP FCM as described. B cells were incubated with lentiviral particles at a multiplicity of infection of 3 in RPMI1640 supplemented with 10% FCS for 18–24 h and then transferred on MS5 stromal cells for at least 8 days. Transduction efficiencies ranged between 10 and 30%.
Flow cytometry measurements
CLL-BC were identified by anti-CD5-APC-Cy7 and anti-CD19-PC7 antibodies. Apoptosis was assessed by Annexin V labeling. MTP measurements were achieved using 3.3′dihexyloxacarbocyanine iodide (Di0C6(3)) or TMRM fluorescent probes, sequestered by active mitochondria.37 Dead cells were stained by 7-AAD or TO-PRO-3 iodide (Life Technologies, Carlsbad, CA, USA). ROS were evaluated using the DHE fluorescent probe that binds to superoxide anions. For STAT3 and pMEK labeling, CD5+CD19+ pre-labeled B cells were fixed, permeabilized and incubated with relevant antibodies, according to Irish et al.38 The antibodies were total STAT3, pSTAT3Tyr705, pSTAT3Ser727 and pMEK (BD Biosciences, San Jose, CA, USA). pSTAT3-Ser727 antibodies were pre-mixed or not with pSTAT3-Ser727 ipep (Santa Cruz, Dallas, TX, USA) or a chemically synthesized STAT3 peptide (IDLPMS727PRTLD) that was phosphorylated (pS3-11pep) or not (S3-11pep) on Ser727 (Thermo Fisher Scientific, Waltham, MA, USA) at a 1 : 100 molar ratio. FCM analyses were performed using Diva and Cellquest on a FACS-Canto II (BD Biosciences). Data are expressed as the percentage of labeled cells among total or as the mean fluorescence intensity.
Immunofluorescence microscopy
B cells, pre-incubated or not for 15 min at 37 °C with red Mitotracker (500 nM, Life Technologies) were loaded on poly-L-lysine-coated slides, fixed in 3.7% paraformaldehyde, permeabilized with phosphate-buffered saline (PBS)/0.1% Triton X-100 for 5 min at room temperature and incubated in 1 × PBS/5% bovine serum albumin (BSA) for 1 h at 37 °C in the presence of primary antibodies. The latter were murine anti-pSTAT3Ser727 antibodies (BD Biosciences), rabbit anti-pSTAT3Ser727 and anti-total STAT3 antibodies (Cell Signaling Technology, Danvers, MA, USA), and antibodies directed to the Mt DNA-encoded complex I (NADH-ubiquinone reductase) subunit (mtND1, gift of Dr. Lombes).16 Rabbit pSTAT3-Ser727 antibodies were pre-mixed or not with the indicated pSTAT3-Ser727 peptide (Santa Cruz) at a 1 : 1 concentration ratio. Then, FITC-conjugated anti rabbit immunoglobulin G antibody (BD Biosciences) or PE-conjugated anti-murine immunoglobulin G (Life Technologies) secondary antibodies were used for 1 h incubation at room temperature. Slides were DAPI-stained and viewed using a Leica confocal microscope (LEICA,Wetzlar, Germany), with a digital AxioCamERc camera and analyzed with AxioVisio software (Carl Zeiss, Jena, Germany). At least 25 cells/patient sample were observed at × 63 and × 100 magnifications. Co-localization analyses used a cooled Coolsnap camera (Roper Scientific, Trenton, NJ, USA) with optical z-sections collected at 0.63 μm steps. Image analyses were performed with the software ImageJ associated with the JACoP plugin tool to integrate statistics to spatial exploration of correlated pixel signals (expressed as Pearson’s correlation coefficient). Coefficients >0.5 indicated significant colocalization.39
Transmission electron microscopy
B cells were fixed in 3% glutaraldehyde and postfixed in 1 × PBS/1% osmium tetroxide, dehydrated in ethanol followed by propylene oxide (VWR, Radnor, PA, USA), embedded in Epon and polymerized overnight at 60 °C. Ultrathin sections were cut with a LEICA/Reichert Ultracut S ultramicrotome, stained with uranyl acetate and lead citrate. Immunolabeling was performed on B cells fixed in 0.2% glutaraldehyde/4% paraformaldehyde for 1 h at room temperature as described,40 using rabbit total STAT3 or pSTAT3Ser727 antibodies (Cell Signaling Technology). Ultrathin (Peabody, MA, USA) sections were examined with a JEOL1011 transmission electron microscope. Digital images of ≥10 cells/sample were obtained using a GATAN-CCD camera and Digital Micrograph software (Warrendale, MA, USA).
Preparation of mitochondria extracts
Mitochondria were isolated from B cells using Mitochondria isolation kit (Miltenyi Biotec) following manufacturer’s instructions with some modifications. Briefly, frozen B cells (2–10 × 107 cells) were directly resuspended in pre-cooled phosphate-buffered saline (1 ml per 108 total cells) supplemented with ethylenediamine tetraacetatic acid (EDTA; 2 mM), anti-protease and –phosphatase inhibitors and benzonase (50 U), and incubated for 20 min at 4 °C. Cell homogenization was performed with a 26-G needle stepwise using 5–10 stokes. Lysates were diluted to 1 × separation buffer (Miltenyi Biotec) and proceeded to magnetic labeling by incubation with anti-Tom22 magnetic beads for 1 h at 4 °C on a wheel. The labeled cell lysate was loaded on a MACS column placed in a magnetic field and let run through. Lysates were re-loaded three times. Column was then intensively washed out and the magnetically labeled mitochondria were then flushed out. Pre- and post-mitochondria-purified fractions were separated by SDS-PAGE and the presence of cytoplasmic β-actin protein, Mt hsp60 or NDUFA9 proteins, and nuclear KDM1a were evaluated by western blot analysis using monoclonal anti-β-actin (AC-15, Sigma-Aldrich), -hsp60 (Biosciences Inc., Allentown, PA, USA), -NDUFA9 (Abcam, Cambridge, UK) and KDM1a (Cell Signaling Technology).
Immunoprecipitation
Total cell extracts from B cells (5 107) were obtained by a 30-min incubation at 4 °C in lysis buffer (20 mM Tris, pH 8, 137 mM NaCl, 1% NP40, 10% glycerol and 2 mM EDTA) supplemented with cocktails of proteinase (Roche) and phosphatase inhibitors. Lysates were immunoprecipitated for 4 h at 4 °C with 4 μg of complex I immunocapture monoclonal antibody (MitoSciences, Eurogen, OR, USA) or isotype-matched control (ctl). Immunoprecipitates were separated by SDS-PAGE and immunoblotted with the indicated antibodies.
Oligo pull-down assays
Cytoplasmic and nuclear extracts were obtained from B cells, as previously described.41 To confirm proper subcellular fractionation, extracts were separated by SDS-PAGE and the presence of the nuclear lamin B and A/C, or the cytoplasmic β-actin was assessed by western blot analysis using monoclonal mouse anti-β-actin (Sigma-Aldrich), polyclonal rabbit anti-lamin B, -lamin A/C (Cell Signaling Technology) antibodies. Nuclear extracts (2–5 × 107) were incubated with 1 μg of the indicated biotin-labeled STAT3-DNA probe SIE367 (5′-TTCCCGTAA-3′) or IRF1 (5′-GATCCATTTCCCCGAAATGA-3′) or control Oct-DNA probe (5′-CGGGTATAATTTCTGC-3′) in binding buffer (10 mM HEPES, 100 μM EDTA, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 4 mM spermidine, 2 mM dithiothreitol, 0.1 mg of BSA/ml, 2.5% glycerol and 4% Ficoll) for 2 h at 4 °C, as described.41 Streptavidin magnetic beads (Pierce, Thermo Fisher Scientific) were added for 30 min at 4 °C, extensively washed, resuspended in SDS-PAGE loading buffer and analyzed by SDS-PAGE and immunoblotting.
Statistics
Data were expressed as means or medians depending on the normality of the distribution. Statistical significance was determined by Student’s t-test or Mann–Whitney/Wilcoxon test. P-values≤0.05 were considered as significant (*) and P-values ≤0.01 as highly significant (**). Statistical analyses were performed with the Medcalc (Ostende, Belgium) and GraphPad Prism softwares (San Diego, CA, USA).
Change history
29 January 2015
This article has been corrected since Online Publication and a corrigendum has also been published
Abbreviations
- CLL:
-
chronic lymphoid leukemia
- CLL-BC:
-
CLL B lymphoid cells
- N-BC:
-
normal B lymphoid cells from healthy donors
- S3:
-
STAT3
- pSTAT3Ser727:
-
Ser727-phosphorylated STAT3
- pSTAT3Tyr705:
-
Tyr705-phosphorylated STAT3
- MTP:
-
mitochondria transmembrane potential
- FCM:
-
flow cytometry measurement
- 7-AAD:
-
7-aminoactinomycin D
- TMRM:
-
tetramethylrhodamine-methyl-ester
- Mt:
-
mitochondrial
- ND1:
-
NADH-ubiquinone reductase subunit
- DHE:
-
dihydroethidium
- PEITC:
-
β-phenylethyl isothiocyanate
- NAC:
-
N-acetyl cysteine
- GSH:
-
glutathione
References
Burger JA . Targeting the microenvironment in chronic lymphocytic leukemia is changing the therapeutic landscape. Curr Opin Oncol 2012; 24: 643–649.
Chiorazzi N, Ferrarini M . B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol 2003; 21: 841–894.
Buchner M, Baer C, Prinz G, Dierks C, Burger M, Zenz T et al. Spleen tyrosine kinase inhibition prevents chemokine- and integrin-mediated stromal protective effects in chronic lymphocytic leukemia. Blood 2010; 115: 4497–4506.
de Totero D, Meazza R, Capaia M, Fabbi M, Azzarone B, Balleari E et al. The opposite effects of IL-15 and IL-21 on CLL B cells correlate with differential activation of the JAK/STAT and ERK1/2 pathways. Blood 2008; 111: 517–524.
Till KJ, Spiller DG, Harris RJ, Chen H, Zuzel M, CLL, Cawley JC . but not normal, B cells are dependent on autocrine VEGF and alpha4beta1 integrin for chemokine-induced motility on and through endothelium. Blood 2005; 105: 4813–4819.
Zhang W, Trachootham D, Liu J, Chen G, Pelicano H, Garcia-Prieto C et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol 2012; 14: 276–286.
Levy DE, Darnell JE Jr . Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 2002; 3: 651–662.
Benekli M, Xia Z, Donohue KA, Ford LA, Pixley LA, Baer MR et al. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood 2002; 99: 252–257.
Turkson J, Jove R . STAT proteins: novel molecular targets for cancer drug discovery. Oncogene 2000; 19: 6613–6626.
Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR . Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev 2007; 21: 1396–1408.
Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE . Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 2009; 324: 1713–1716.
Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T et al. Function of mitochondrial Stat3 in cellular respiration. Science 2009; 323: 793–797.
Frank DA, Mahajan S, Ritz J . B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest 1997; 100: 3140–3148.
Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X et al. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood 2010; 115: 2852–2863.
Yang J, Stark GR . Roles of unphosphorylated STATs in signaling. Cell Res 2008; 18: 443–451.
Procaccio V, Mousson B, Beugnot R, Duborjal H, Feillet F, Putet G et al. Nuclear DNA origin of mitochondrial complex I deficiency in fatal infantile lactic acidosis evidenced by transnuclear complementation of cultured fibroblasts. J Clin Invest 1999; 104: 83–92.
Kurtova AV, Balakrishnan K, Chen R, Ding W, Schnabl S, Quiroga MP et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood 2009; 114: 4441–4450.
Carew JS, Nawrocki ST, Xu RH, Dunner K, McConkey DJ, Wierda WG et al. Increased mitochondrial biogenesis in primary leukemia cells: the role of endogenous nitric oxide and impact on sensitivity to fludarabine. Leukemia 2004; 18: 1934–1940.
Armann B, Hanson MS, Hatch E, Steffen A, Fernandez LA . Quantification of basal and stimulated ROS levels as predictors of islet potency and function. Am J Transplant 2007; 7: 38–47.
Mi L, Di Pasqua AJ, Chung FL . Proteins as binding targets of isothiocyanates in cancer prevention. Carcinogenesis 2011; 32: 1405–1413.
Gough DJ, Koetz L, Levy DE . The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLoS One 2013; 8: e83395.
Frecha C, Costa C, Levy C, Negre D, Russell SJ, Maisner A et al. Efficient and stable transduction of resting B lymphocytes and primary chronic lymphocyte leukemia cells using measles virus gp displaying lentiviral vectors. Blood 2009; 114: 3173–3180.
Lee YK, Shanafelt TD, Bone ND, Strege AK, Jelinek DF, Kay NE . VEGF receptors on chronic lymphocytic leukemia (CLL) B cells interact with STAT 1 and 3: implication for apoptosis resistance. Leukemia 2005; 19: 513–523.
Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 2006; 442: 823–826.
Ng YP, Cheung ZH, Ip NY . STAT3 as a downstream mediator of Trk signaling and functions. J Biol Chem 2006; 281: 15636–15644.
Ohta S, Misawa A, Fukaya R, Inoue S, Kanemura Y, Okano H et al. Macrophage migration inhibitory factor (MIF) promotes cell survival and proliferation of neural stem/progenitor cells. J Cell Sci 2012; 125 (Pt 13): 3210–3220.
Courapied S, Sellier H, de Carne Trecesson S, Vigneron A, Bernard AC, Gamelin E et al. The cdk5 kinase regulates the STAT3 transcription factor to prevent DNA damage upon topoisomerase I inhibition. J Biol Chem 2010; 285: 26765–26778.
Szczepanek K, Chen Q, Derecka M, Salloum FN, Zhang Q, Szelag M et al. Mitochondrial-targeted Signal transducer and activator of transcription 3 (STAT3) protects against ischemia-induced changes in the electron transport chain and the generation of reactive oxygen species. J Biol Chem 2011; 286: 29610–29620.
Zhou L, Too HP . Mitochondrial localized STAT3 is involved in NGF induced neurite outgrowth. PLoS One 2011; 6: e21680.
Zhang Q, Raje V, Yakovlev VA, Yacoub A, Szczepanek K, Meier J et al. Mitochondrial localized Stat3 promotes breast cancer growth via phosphorylation of serine 727. J Biol Chem 2013; 288: 31280–31288.
Oltra AM, Carbonell F, Tormos C, Iradi A, Saez GT . Antioxidant enzyme activities and the production of MDA and 8-oxo-dG in chronic lymphocytic leukemia. Free Radic Biol Med 2001; 30: 1286–1292.
Zhou Y, Hileman EO, Plunkett W, Keating MJ, Huang P . Free radical stress in chronic lymphocytic leukemia cells and its role in cellular sensitivity to ROS-generating anticancer agents. Blood 2003; 101: 4098–4104.
Mari M, Morales A, Colell A, Garcia-Ruiz C, Kaplowitz N, Fernandez-Checa JC . Mitochondrial glutathione: features, regulation and role in disease. Biochim Biophys Acta 2013; 1830: 3317–3328.
Shaw PE . Could STAT3 provide a link between respiration and cell cycle progression? Cell Cycle 2010; 9: 4294–4296.
Xie Y, Kole S, Precht P, Pazin MJ, Bernier M . S-glutathionylation impairs signal transducer and activator of transcription 3 activation and signaling. Endocrinology 2009; 150: 1122–1131.
Phillips D, Reilley MJ, Aponte AM, Wang G, Boja E, Gucek M et al. Stoichiometry of STAT3 and mitochondrial proteins: Implications for the regulation of oxidative phosphorylation by protein-protein interactions. J Biol Chem 2010; 285: 23532–23536.
Cottet-Rousselle C, Ronot X, Leverve X, Mayol JF . Cytometric assessment of mitochondria using fluorescent probes. Cytometry A 2011; 79: 405–425.
Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 2004; 118: 217–228.
Bolte S, Cordelieres FP . A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 2006; 224 (Pt 3): 213–232.
Berger G, Masse JM, Cramer EM . Alpha-granule membrane mirrors the platelet plasma membrane and contains the glycoproteins Ib, IX, and V. Blood 1996; 87: 1385–1395.
Magne S, Caron S, Charon M, Rouyez MC, STAT5 Dusanter-Fourt I . and Oct-1 form a stable complex that modulates cyclin D1 expression. Mol Cell Biol 2003; 23: 8934–8945.
Acknowledgements
We thank D Borgel, E Lauret, A Lombès, F Bouillaud and S Martin-Lannerée for their helpful discussions or assistance, Dr. Jacqueline Roussi, Lise Willems and Didier Bouscary for patient samples and BD Biosciences for antibodies. This work received a financial support from INSERM, AP-HP, LNCC (I.D-F) and ARC (I.D-F).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Diederich
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International Licence. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons licence, users will need to obtain permission from the licence holder to reproduce the material. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0
About this article
Cite this article
Capron, C., Jondeau, K., Casetti, L. et al. Viability and stress protection of chronic lymphoid leukemia cells involves overactivation of mitochondrial phosphoSTAT3Ser727. Cell Death Dis 5, e1451 (2014). https://doi.org/10.1038/cddis.2014.393
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.393
This article is cited by
-
STAT3 mediates C6-ceramide-induced cell death in chronic lymphocytic leukemia
Signal Transduction and Targeted Therapy (2017)