Abstract
Background:
Triple-negative breast cancer (TNBC) accounts for 15–20% of all breast cancer in women globally. This subtype often has early and high recurrence rates, resulting in poor survival, partially due to lack of targeted therapies. To date, the detailed molecular mechanisms underlying TNBC progression are unclear. Given the crucial role of microRNAs (miRNAs) in cancer metastasis, we aimed to analyse the expression and function of a metastasis-associated miRNA named miR-211-5p in TNBC.
Methods:
MiRNA array analysis was performed to search for metastasis-associated miRNAs in TNBC. The miR-211-5p expression in tumour tissues, adjacent non-tumourous breast tissues of TNBC patients and cell lines were evaluated by real-time PCR. The protein expression levels were analysed by western blot, immunohistochemistry and in situ hybridisation. Luciferase reporter assays were employed to validate the target of miR-211-5p. The effect of miR-211-5p on TNBC progression was investigated in vitro and in vivo.
Results:
MiR-211-5p was significantly downregulated in TNBC, and its expression level was associated with overall survival in TNBC. The expression of miR-211-5p suppressed TNBC cell proliferation, invasion, migration and metastasis in vitro and in vivo. Furthermore, SETBP1 was identified as a target of miR-211-5p. Through gain-of-function and loss-of-function studies, SETBP1 was shown to significantly affect colony and cell number in vitro. Enforced expression of miR-211-5p inhibited the expression of SETBP1 significantly and the restoration of SETBP1 expression reversed the inhibitory effects of miR-211-5p on TNBC cell proliferation and metastasis.
Conclusions:
These findings collectively demonstrate a tumour suppressor role of miR-211-5p in TNBC progression by targeting SETBP1, suggesting that miR-211-5p could serve as a potential prognostic biomarker and therapeutic target for TNBC.
Similar content being viewed by others
Main
Breast cancer, the most common malignancy in women, is a heterogeneous disease with substantial diversity in histological and molecular characteristics, which can be divided into four major molecular subtypes: luminal A, luminal B, triple-negative/basal-like and HER2 type (Prat et al, 2014; Shin et al, 2015; Woolston, 2015; Yaffe and Jong, 2016; Braunstein et al, 2017). Of them, triple-negative breast cancers (TNBC) represent ∼15–20% of all breast cancers and is characterised by lack of oestrogen receptor (ER), progesterone receptor and human epidermal growth factor receptor 2 (HER2). TNBC highly express epidermal growth factor receptor-1 (EGFR) and are highly proliferative, aggressive and resistant to systemic chemotherapies (Bauer et al, 2007; Boyle, 2012). Consequently, the outcome for these patients is poor compared with ER+ and HER2+ cancers. Approximately 30–40% of deaths are caused by recurrence and metastasis of TNBC. Even though EGFR inhibitors have shown some promise for treating TNBC, no FDA-approved targeted therapies have improved patient outcome for TNBC (Dent et al, 2007; Foulkes et al, 2010; Kwilas et al, 2016; Shu et al, 2016; van Roozendaal et al, 2016). Therefore, a better understanding of the molecular events underlying the metastasis of TNBC is important for its prevention, diagnosis and treatment.
MicroRNAs (miRNAs) are highly conserved small non-coding regulatory RNAs with sizes of 17–25 nucleotides. As posttranscriptional regulators, miRNAs can negatively regulate gene expression by binding directly to the 3′ untranslated region (3′-UTR) of corresponding target messenger RNAs (mRNAs) in a sequence-specific manner, which induces mRNA degradation or protein translation repression (Ambros, 2004; Bartel, 2004; Elmén et al, 2008; Kota et al, 2009; Yates et al, 2013). Mounting evidence links miRNAs to the initiation, promotion and progression of cancer, and miRNAs play critical roles in pathological processes of breast cancer, such as cell proliferation, metastasis, migration and DNA methylation (Ma et al, 2007; Dvinge et al, 2013; Verghese et al, 2013; Kim et al, 2016). Although the importance of miRNAs in metastasis has attracted much attention in recent years, the pathological relevance and significance of the majority of miRNAs in TNBC remain unclear.
In this study, using miRNA array, we found that miR-211-5p was downregulated in TNBC and that its expression levels were associated with overall survival in breast cancer. Furthermore, miR-211-5p inhibited TNBC cell proliferation, invasion, migration and metastasis in vitro and in vivo by targeting SETBP1. Enforced expression of miR-211-5p inhibited the expression of SETBP1 significantly and the restoration of SETBP1 expression reversed the inhibitory effects of miR-211-5p on TNBC cell proliferation and metastasis. These findings show that miR-211-5p functions as a tumour suppressor in TNBC development, which could provide a promising prognostic and therapeutic strategy for TNBC treatment.
Materials and methods
Clinical samples
Between March 2011 and September 2015, tissue samples including 82 triple-negative breast cancer tissues (TNBC), 68 corresponding normal adjacent tissues, 98 non-triple-negative breast cancer tissues (NTNBC) and 87 corresponding normal adjacent tissues were obtained from the routine therapeutic surgery of patients. In addition, 82 serum samples from TNBC patients and 68 serum samples from normal controls were also obtained. All these patients did not receive any anti-tumour treatment and all specimens were confirmed by pathology. The pathological data of TNBC were described in Supplementary Table 1. This study protocol was approved by the Ethics Committee of our institution, and written informed consent was obtained from each participant.
Cell lines and transfection
The following cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA), that is, MDA-MB-231, MDAMB-361, MDA-MB-435, MDA-MB-468, MCF-7, T47D, BT-474, BT-20 and BT-483 and normal mammary epithelial cell lines (HBL-100, 184A and MCF-10A). Cells were cultured in RPMI 1640 (Gibco-BRL, Karlsruhe, Germany), supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL) and 100 U ml−1 of penicillin and streptomycin (Gibco-BRL), and maintained at 37 °C with 5% CO2. All transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. MiR-211-5p mimic, miR-211-5p inhibitor and their related negative control (Applied Biosystems, Foster City, CA, USA) were transfected into cells at a concentration of 50 nM, and the cells were maintained 48 h after transfection. Constructs containing shSETBP1 and its negative control shRNA, pcDNA3.1-SETBP1 and its vector pcDNA3.1 were used. The constructs were transfected into cells at a concentration of 4 μg, and selected by neomycin at 48 h after transfection.
Lentivirus preparation
Lentiviral vector expressing pre-miR-211-5p plasmid (lenti.miR-211-5p) and lenti.GFP (control) were generated by removal of GFP from pRRLsinpptCMV.GFP.WPRE by BamHI/SalI digestion and ligation of pre-miR-211-5p sequence. Lenti.GFP served as control. Lentiviral vectors were produced by co-transfection of cell and the virus harvested by ultracentrifugation of the supernatant. Viral p24 concentration was determined by immunocapture (Alliance, Perkin Elmer Life Sciences Inc., Anaheim, CA, USA) and transducing activity quantified. Cells were plated 24 h prior to infection, incubated with fresh media containing the required multiplicity of infection per cell (m.o.i.) of virus, left for 1 h, washed and maintained until harvesting. All lentivirus infections were performed in the presence of 8 μg ml−1 polybrene (Sigma, Poole, UK).
RNA isolation
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) and RNeasy mini kit (Qiagen, Valencia, CA, USA) according to the manufacturers’ protocol. Subsequently, RNA was eluted in 50 μl of nuclease-free water, and stored at −80 °C for further analysis. The RNA concentration was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Microarray analysis
Ten TNBC tissues and 10 corresponding normal adjacent tissues (also serum samples) were randomly selected for miRNA array analysis. Samples were prepared according to Affymetrix protocols (Affymetrix, Santa Clara, CA, USA). RNA quality and quantity was ensured using the Bioanalyzer (Agilent, Santa Clara, CA, USA) and NanoDrop (Thermo Scientific, Waltham, MA, USA), respectively. Per RNA labelling, 500 ng of total RNA was used in conjunction with the Affymetrix recommended protocol for the HG_U133_Plus 2.0 chips. The hybridisation cocktail containing the fragmented and labelled cDNAs was hybridised to the Affymetrix Human HG_U133_Plus 2.0 GeneChip. The chips were washed and stained by the Affymetrix Fluidics Station using the standard format and protocols as described by Affymetrix and the Affymetrix GeneChip Scanner 3000 was used to scan the probe arrays. Gene expression intensities were extracted using Affymetrix AGCC software. Partek Genomic Suite was used to RMA normalise (Robust Multichip Analysis), summarise, log2 transform the data and run the ANOVA analysis.
Quantitative RT–PCR analysis
Reverse transcription (RT) and quantitative RT–PCR (qRT–PCR) reactions were performed by means of a qSYBR-green-containing PCR kit (Qiagen, Germantown, MD, USA). The fold change was determined as 2−ddCt, where Ct is the fractional cycle number at which the fluorescence of each sample passes the fixed threshold. The dCt was calculated by subtracting the Ct of snRNA U6 from the Ct of the miRNA of interest. The ddCt was calculated by subtracting the dCt of the reference sample (paired non-tumourous tissue for the surgical samples, and MCF-10 cells for the nine breast cancer cell lines) from the dCt of each sample. The sequences of the specific primers for miR-211-5p and U6 snRNA were 5′-GATGCTGTAATGGATGATATGA-3′ and 5′-ATTGGAACGATACAGAGAAGATT-3′, respectively. The primers for the qRT–PCR detection of SETBP1 mRNA were synthesised by Invitrogen: forward, 5′-CAGAAGCCCAAACCAAAA-3′ and reverse, 5′-TTCAGTCAGCACCTTATCAG-3′. All of the real-time PCR assays were performed with the Bio-Rad (Hercules, CA, USA) IQTM5 Multicolour Real-Time PCR Detection System (USA).
Luciferase constructs and reporter assay
For construction of the wild-type SETBP1 3′UTR-Luc reporter plasmid, a fragment of the 3′UTR of the SETBP1 gene, including the predicted miR-211-5p-binding site, was PCR amplified using the primer sets: forward 5′-CTCGAGTAACCGTATTGTTCGCGTCATGCCAGCA-3′ and reverse 5′-GCGGCCGCCAGACCATGTGTCCCATTTG-3′, and then cloned into psi-CHECKTM-2 vector downstream of Firefly Luciferase with XhoI and NotI. To produce constructs that bear mutations at a putative miR-211-5p-binding site in wild-type SETBP1 3′UTR, site-directed mutagenesis was performed. The wild-type SETBP1 3′UTR constructs were mutated with the following oligonucleotide primer sets: forward 5′-GGTTCTGTGAACAATTGGCAG TAAGTTATC-3′ and reverse 5′-GATAACTTACTGCCAATTGTTCACAGAACC-3′. The PCR mixture had 0.7 μl of expand long-range enzyme mix (Roche, Mannheim, Germany), 10 μl of 5 × expand long-range buffer, 100 ng of plasmid template, 100 nM of primers, 3 μl of DMSO and 2.5 μl of dNTPs (10 mM). PCR cycling conditions were as follows: 92 °C for 30 s, 55 °C for 1 min, 68 °C for 10 min and a final extension at 68 °C for 10 min. After PCR, 20 μl of the reaction was digested with DpnI at 37 °C for 1 h, and 10 μl was transformed into DH5 alpha E. coli to prepare the mutant construct plasmids. All constructs were confirmed by sequencing (Cosmogenetech). Cells were transfected with combinations of wild type or mutant-type SETBP1 3′UTR-Luc reporter plasmid and mimic control (Applied Biosystems), miR-211-5p mimic (Applied Biosystems), inhibitor control (Applied Biosystems) and miR-211-5p inhibitor (Applied Biosystems) using Lipofectamine 2000. Forty-eight hours after transfection, luciferase activity was determined with a Dual-Glo Luciferase Assay system (Promega, Madison, WI, USA), according to the manufacturer’s instructions. All assays were performed in triplicate.
Colony formation assays
Cells were suspended in DMEM containing 0.35% low-melting agarose and plated onto solidified 0.6% agarose containing DMEM in six-well culture plates at a density of 1 × 105 cells per dish. The plates were incubated for 2 weeks at 37 °C in a 5% CO2 incubator, and the number of colonies was counted after staining with 0.1% crystal violet solution.
Flow cytometry
Apoptosis was evaluated by staining cells with both Annexin V-FITC and PI, according to the manufacturer’s instructions. Annexin V-FITC was employed to quantitatively determine the percentage of cells undergoing apoptosis. It relies on the property of cells to lose membrane asymmetry in the early phase of apoptosis. In apoptotic cells, the membrane phospholipid phosphatidylserine is translocated from the inner leaflet of the plasma membrane to the outer leaflet, thereby exposing phosphatidylserine to the external environment. Cells that were positively stained with Annexin V-FITC and negatively stained for PI were considered apoptosis. Cells that were positively stained for both Annexin V-FITC and PI were considered necrosis. To quantitate apoptosis, the cells were washed with cold phosphate-buffered saline solution and then resuspended in binding buffer (10 mmol l−1 HEPES (N-2-hydroxyethylpiperazine-N‘-2-ethanesulphonic acid)/NaOH (pH 7.4), 140 mmol l−1 NaCl and 2.5 mmol l−1 CaCl2). The cells were stained with 5 μl Annexin V-FITC and 10 μl PI and then analysed with EpicsAltra (Beckman Coulter, CA, USA) FCM.
TUNEL assays
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling (TUNEL) staining kit (Roche) was used to analyse the cell apoptosis, by following the manufacturer’s instructions.
Cell proliferation assay
MDA-MB-231 and MDA-MB-435 cells were seeded in 24-well plates overnight and then transfected with blank, miR-211-5p mimic and miR-211-5p inhibitor) (final concentration 200 nM). Afterwards, cells were trypsinised and counted, seeded in 96-well plates (in triplicate) for an MTT assay at a density of 8000 cells per well (MDA-MB-231) or 10 000 cells per well (MDA-MB-435), and incubated at 37 °C for 24 h. Then, at 24, 48 and 72 h after cell seeding, 10 μl MTT (0.5 mg ml−1; Sigma-Aldrich, St Louis., MO, USA) was added to each well (20 μl per well), and the cells were maintained at 37 °C for another 4 h. The medium was removed, and the precipitate was dissolved in 100 μl DMSO (Sigma). After shaking for 15 min, the absorbance at 570 nm (A570) was measured using an ELISA reader at a wavelength of 570 nm.
Transwell invasion and migration assay
Invasion assay was performed with the Transwell chamber with 8 μm pores (Corning, Tewksbury, MA, USA). Fifty microliters diluted matrigel (2 mg ml−1, BD Biosciences, Bedford, MA, USA) was placed on the inner surface. MDA-MB-231 and MDA-MB-435 cells were transfected for 24 h and isolated to make a final concentration at 2 × 105 ml−1, which then placed on the top chamber. RMPI1640 with 20% FBS was added to the bottom chamber. After 24 h, non-invading cells were removed from the top of the matrigel with a cotton-tipped swab. Invading cells at the bottom of the matrigel were fixed in methanol and stained with crystal violet. The invasiveness was determined by counting the penetrated cells under a microscope at × 200 magnification of five random fields in each well. Each experiment was performed in triplicate. For the transwell migration assay, the process was similar with the transwell invasion assay except the inner surface of the chamber without matrigel.
EdU assay
Cell proliferation was measured using EdU kits (RIBOBIO, Shanghai, China) according to the manufacturer’s instructions to analyse the incorporation of EdU during DNA synthesis. Assays were performed in triplicate.
Wound-healing migration assay
MDA-MB-231 and MDA-MB-435 cells were transfected for 24 h, and isolated to make a final concentration at 2 × 105 ml−1, which then plated in 12-well plates (2 × 105 per well) for 24 h. When the cells reached 90% confluence, sterile pipette tips were used to scratch the wound uniformly. Cell motility was assessed by measuring the movement of cells into a scraped wound. The speed of wound closure was monitored after 72 h by measuring the distance of the wound from 0 h. Each experiment was conducted in triplicate.
In situ hybridisation analysis and immunohistochemistry analysis
In situ hybridisation (ISH) procedures were carried out as previously described. MiR-211-5p miRCURYTM LNA custom detection probes (Exiqon, Vedbaek, Denmark) were used for ISH. The 5′–3′ sequence (enhanced with LNA) was UACAGUACUGUGAUAACUGAA with digoxigenin at the 5′ and 3′ ends. Hybridisation, washing and scanning were performed according to the manufacturer’s instructions. Staining intensity was scored as 0 (negative), 1+ (weak), 2+ (medium) or 3+ (strong). Low expression was defined as a staining intensity of 0, 1, 2 or 3 with <10% of cells being stained or an intensity of 0 or 1 with <50% of cells stained. High expression was defined as an intensity of 2 or 3 with <10% of cells stained or an intensity of 1, 2 or 3 with <50% of cells. Immunohistochemistry was performed on tissue microarray sections using anti-SETBP1 antibody (Santa Cruz, CA, USA). The complex was visualised with streptavidin/peroxidase and DAB complex, and the nuclei were counterstained with haematoxylin. The intensities of miR-211-5p and SETBP1 staining were scored between 0 and 4 according to a previous description.
Western blot
Protein was extracted from TNBC cell lines using RIPA lysis buffer with a proteinase inhibitor. The protein concentration in the lysates was measured with the Protein BCA Assay Kit (Bio-Rad), and 20 μg of protein mixed with 2 × SDS loading buffer was loaded per lane. The proteins in the lysates were separated by 12% SDS polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore Corporation, Billerica, MA, USA). To block nonspecific binding, the membranes were incubated at room temperature for 1 h with 5% skim milk powder. The membranes were then incubated for 12 h at 4 °C with an antiserum containing antibodies against SETBP1 purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). A peroxidase-conjugated secondary antibody and ECL western blotting detection reagents were used to visualise the target proteins (ECL Millipore Corporation), which were quantified with a Bio Image Intelligent Quantifier 1-D (Version 2.2.1, Nihon-BioImage Ltd., Japan). An anti-GAPDH antibody (1 : 5000, Abcam, Cambridge, UK) was used as a protein loading control.
Mouse xenograft model
The breast cancer model in nude mice was constructed as previously described. A total of 2 × 106 MDA-MB-231 cells infected with miR-211-5p or scramble viruses were propagated and inoculated subcutaneously into the dorsal flanks of nude mice (five in each group). The tumour size was measured every 4 days. After 28 days, the mice were killed, necropsies were performed and the tumours were weighed. The tumour volumes were determined according to the following formula: A × B2/2, where A is the largest diameter and B is the diameter perpendicular to A.
To determine the effect of miR-211-5p on TNBC metastatic behaviour in vivo, we performed a mouse orthotopic mammary xenograft tumour model study by injecting 4T1 or miR-211-5p overexpression 4T1 cells into the mammary fat pad (eight in each group). The bioluminescence procedure was performed. The numbers of micrometastases in the lung from spontaneous metastasis after orthotopic injection of 4T1 cells were analysed by morphological observation. Cell proliferation (Ki67) was also evaluated. All of the animal procedures were carried out with ethical committee approval and meet the standards required by the UKCCCR guidelines (Workman et al, 2010).
Statistical analysis
The statistical analyses were performed using SPSS 17.0 software (SPSS Inc, Chicago, IL, USA), and graphs were generated using GraphPad Prism 5 Software (GraphPad Software, Inc, La Jolla, CA, USA). Paired t-test, Student’s t-test and Kruskal–Wallis tests were used to analyse miRNA and gene expression. An ANOVA was also performed comparing more than two groups. The Pearson’s correlation test was employed to evaluate the associations. Overall survival curves and relapse-free curves were plotted according to the Kaplan–Meier method, and the log-rank test was used for comparison. Survival was counted from the day of the surgery. All of the differences were statistically significant at the P<0.05 level.
Results
Identification of miRNAs differentially expressed in TNBC tissues
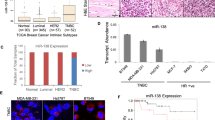
We identified 14 289 miRNAs expressed in TNBC tissues and 2107 were upregulated and 1623 were downregulated significantly when compared with normal adjacent tissues (fold change ⩾2, P value <0.05) (Figure 1A). These differentially expressed miRNAs were chosen for further study only when they met the following criteria: (1) having at least 20 copies of miRNA expression; (2) mean fold change >6 or <0.2; and (3) P values <0.05. On the basis of these criteria, 18 miRNAs, of which 11 miRNAs were upregulated and 7 miRNAs were downregulated in TNBC patients compared with controls, were chosen for further validation (Figure 1B). Also, the miRNA expression profile in serum from TNBC patients were presented (Figure 1C), which was consistent with that in TNBC tissues.
The miR-211-5p expression level was downregulated in TNBC tissues. (A) Volcano plot of the differentially expressed miRNAs. The red points in the plot represent the differentially expressed miRNAs with statistical significance. (B) Heat map was generated by unsupervised clustering analysis with 18 significantly dysregulated miRNAs in TNBC tissues. Hierarchical clustering was performed with average linkage and uncentred correlation. (C) MiRNA expression profile in serum samples from TNBC patients. (D) Expression levels of miR-211-5p determined by qRT–PCR in three mammary normal cell lines and tumour cell lines. The miR-211-5p expression was normalised based on the expression of U6 RNA (six replicates per group, three independent experiments per group). (E) Expression levels of miR-211-5p in 82 TNBC tissues and 68 corresponding normal adjacent tissues and in 98 NTNBC tissues and 87 corresponding normal adjacent tissues. (F) Low levels of miR-211-5p correlated with shorter survival. Overall survival curves for all of the studied patients with high or low miR-211-5p expression. The results demonstrated that patients with low miR-211-5p expression exhibited shorter mean months of overall survival than patients with high miR-211-5p expression. TNBC, triple-negative breast cancers; NTNBC, non-triple-negative breast cancers; Hsa, human; ***P<0.001. Error bars represent the mean±s.e.m. A full colour version of this figure is available at the British Journal of Cancer journal online.
In the training set, miRNAs were measured in a separate set of samples from 10 TNBC patients and 10 controls of the previous step. Only miRNAs with a mean fold change >6 or <0.2 and a P value <0.01 were selected for further analysis. Using the above-mentioned criteria, miR-188-5p, miR-140 and miR-211-5p were observed to be significantly dysregulated in TNBC patients compared with controls (Table 1).
In the validation set, the concentration of miR-188-5p, miR-140 and miR-211-5p were measured by qRT–PCR in a larger cohort comprising of 82 patients and 68 controls. The miR-211-5p expression pattern in the validation set was consistent with those in the training set. Compared with the controls, the level of miR-211-5p was significantly lower in TNBC patients (Table 1). Thus, we focused on miR-211-5p for further study. The workflow of the study design is shown in Supplementary Figure 1.
MiR-211-5p is downregulated in TNBC
MiR-211-5p was highly expressed in normal mammary cell lines, while its expression level in some tumour cell lines was low, particularly those classified as basal-like (Figure 1D). Then, miR-211-5p was measured in 82 TNBC tissues and 68 matched adjacent tissues and in 98 pairs of NTNBC tissues and 87 matched adjacent tissues. The results demonstrated that the level of miR-211-5p was downregulated in TNBC tissues when compared their matched adjacent tissues (Figure 1E). However, the miR-211-5p level was only slightly reduced in NTNBC tumours. Of note, the miR-211-5p level was significantly reduced in TNBC compared with NTNBC (Figure 1E). In addition, its expression level was associated with overall survival in TNBC (Figure 1F).
MiR-211-5p suppress cell proliferation and induce apoptosis in vitro
To illustrate the function of miR-211-5p in TNBC, miR-211-5p was overexpressed in MDA-MB-231 and MDA-MB-435 cell lines by transfecting miR-211-5p mimic, followed by RT–qPCR (Figure 2A). Further, cell proliferation was measured by the CCK-8 (Figure 2B) and EdU assay (Figure 2C), and miR-211-5p significantly suppressed cell proliferation in MDA-MB-231 and MDA-MB-435 cell lines. Moreover, relative to the NC group, colony formation was inhibited by miR-211-5p in MDA-MB-435 and MDA-MB-231 cells in soft agar assay (Figure 2D). In addition, flow cytometry (FCM) indicated similar cell cycle distribution and apoptosis in cells with knockdown of miR-211-5p through induction of G0/G1 phase arrest (Figure 2E and F). The data collectively indicated that miR-211-5p inhibits cell proliferation and induces apoptosis.
MiR-211-5p inhibited breast cancer progression in vitro. (A) Cells were transfected with mimic control and miR-211-5p mimic, respectively, and 48 h later, the expression of miR-211-5p was determined using RT–qPCR (three replicates per group, three independent experiments per group); cell proliferation was evaluated by the CCK-8 assay at 24, 48 and 72 h (B) (eight replicates per group, three independent experiments per group) and EdU assay (C) Magnification × 400). n=3, ***P<0.001. (D) The effect of miR-211-5p on colony formation was evaluated by the soft agar assay. Magnification × 200. (E) The effects of miR-211-5p on cell cycle was determined by FCM analysis. (F) MiR-211-5p mimic can promote cell apoptosis detected by FCM. (G) Apoptosis was assessed in cells transfected with miR-control or miR-211-5p by TUNEL. Scale bar: 50 um. Error bars represent the mean±s.e.m. A full colour version of this figure is available at the British Journal of Cancer journal online.
MiR-211-5p can suppress breast cancer cells invasion and migration
To further investigate whether miR-211-5p affects cell metastasis, transwell assay and wound-healing assay were performed. Transwell invasion assay with matrigel demonstrated that miR-211-5p mimic can inhibit the invasion of MDA-MB-231 and MDA-MB-435 cells by 76% (Figure 3A), and transwell migration assay without matrigel further showed that miR-211-5p can suppress MDA-MB-231 and MDA-MB-435 cells migration by 89% (Figure 3B). Furthermore, we used wound-healing assay to detect the function of miR-211-5p on cell migration. As shown in Figure 3C, the miR-211-5p mimic inhibited the potential of MDA-MB-231 and MDA-MB-435 cells migration. These results suggested that miR-211-5p can suppress breast cancer cells invasion and migration.
MiR-211-5p suppressed the invasion and migration of breast cancer cells. (A) Transwell invasion assay with matrigel were performed in miR-211-5p mimic or mimic control transfected MDA-MB-435 and MDA-MB-231 cells (six replicates per group, three independent experiments per group). Magnification × 400. (B) Transwell migration assay without matrigel were performed in miR-211-5p mimic or mimic control transfected MDA-MB-231 and MDA-MB-435 cells (six replicates per group, three independent experiments per group). Magnification × 400. (C) Wound-healing assay was performed in MDA-MB-435 and MDA-MB-231 cells transfected with miR-211-5p mimic or mimic control (six replicates per group, three independent experiments per group). ***P<0.001. Error bars represent the mean±s.e.m. A full colour version of this figure is available at the British Journal of Cancer journal online.
The effect of miR-211-5p on tumorigenesis and metastasis in a xenograft model
To directly evaluate the role of miR-211-5p in tumour formation and growth in vivo, the xenograft model of human TNBC cells in nude mice was adopted. Briefly, MDA-MB-231 cells infected with miR-211-5p mimic or mimic control lentivirus were injected subcutaneously into each flank of nude mice. After the cells were injected, the tumour volume was monitored every 4 days, and the growth curves of the tumours were plotted accordingly. Finally, all of the mice were killed to harvest the xenograft. It is obvious that the mean volume and weight of the tumours generated from the miR-211-5p overexpression group was significantly lower compared with the control group (Figure 4A–C). Then, we studied the effect of miR-211-5p on tumour metastasis in vivo. We performed a mouse orthotopic mammary xenograft tumour model study by injecting 4T1 or miR-211-5p overexpression 4T1 cells into the mammary fat pad. The bioluminescence image showed that miR-211-5p can effectively inhibit lung metastasis (Figure 4D). After 8 weeks, the mice were anaesthetised, and their lungs were dissected. Haematoxylin and eosin staining was performed to evaluate the tissue morphology (Figure 4E). As shown in Figure 4E, a significantly lower number of macroscopic lung metastases could be observed in mice infected with miR-211-5p mimic. In addition, miR-211-5p can inhibit tumour cell proliferation (Figure 4F). These results indicate that miR-211-5p may repress TNBC proliferation and metastasis.
MiR-211-5p inhibited TNBC cell growth and metastasis in vivo. (A, B, C) Tumour growth in mouse xenograft models. MDA-MB-231 cells infected with miR-211-5p mimic lentivirus or mimic control were injected subcutaneously into nude mice. The tumour volume was measured every 4 days (three replicates per group, six independent experiments per group). After 28 days, the mice were killed, necropsies were performed and the tumours were weighed (three replicates per group, six independent experiments per group). (D, E, F) Tumour metastasis in a mouse orthotopic mammary xenograft tumour model. 4T1 cell overexpressing miR-211-5p mimic or mimic control were injected into the mammary fat pad. After 8 weeks, the mice were killed. MiR-211-5p can substantially suppress lung metastasis. The micrometastases in the lung per HE-stained section from individual mice were calculated and tumour cell proliferation was evaluated. Each group had eight mice. ***P<0.001. Magnification × 400. A full colour version of this figure is available at the British Journal of Cancer journal online.
Validation of SETBP1 as a direct downstream target of miR-211-5p
To gain further insight into the molecular mechanism of miR-211-5p in the development of TNBC, we sought out to determine its gene targets in the MDA-MB-231 and MDA-MB-435 cells by interrogating the interaction between miR-211-5p and its target mRNA transcripts. As predicted by miRanda (http://www.microrna.org/), TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and PITA (http://genie.weizmann.ac.il/pubs/mir07/), there was complementarity between miR-211-5p and SETBP1 3′UTR (Figure 5A and B). To further confirm the functional interaction between miR-211-5p and SETBP1 generated by the target prediction algorithms, we performed luciferase reporter assays with a SETBP1 vector that contained either the putative miR-211-5p-binding sites (wild type) or the mutant-binding sites (MUT) that were contained in the 3′UTR (Figure 5C). Overexpression of miR-211-5p was achieved by transfecting MDA-MB-231 and MDA-MB-435 cells with a miR-211-5p mimic, while knockdown of miR-211-5p was achieved by transfecting MDA-MB-231 and MDA-MB-435 cells with a miR-211-5p inhibitor. Overexpression of miR-211-5p significantly reduced luciferase activity of the reporter gene in wild type, but not mutant, indicating that miR-211-5p directly targeted the SETBP1 3′UTR (Figure 5D). This effect was further confirmed by gene and protein expression. As shown in Figure 5E and F, overexpression of miR-211-5p decreased both SETBP1 protein and mRNA levels in MDA-MB-231 and MDA-MB-435 cells, while inhibition of miR-211-5p increased SETBP1 level. Taken together, the results demonstrated that miR-211-5p directly recognises the 3′-UTR of SETBP1 transcripts and regulates its expression at the posttranscriptional level.
Identification of SETBP1 as a target of miR-211-5p. (A) Venn diagram displaying miR-211-5p computationally predicted to target SETBP1 by four different prediction algorithms: miRanda, TargetScan, PicTar and PITA. (B) The structure of miR-211-5p. (C) Schematic representation of SETBP1 3′UTR demonstrating putative miRNA target site, luciferase activities of wild-type (WT-UTR) and mutant (MUT-UTR) constructs. (D) Luciferase reporter activity following expression after transfections (mimic control, miR-211-5p mimic, inhibitor control, miR-211-5p inhibitor) in MDA-MB-435 and MDA-MB-231 cells (three replicates per group, six independent experiments per group). (E) Real-time PCR analysis showed that overexpression of miR-211-5p can decrease SETBP1 mRNA level, while inhibition of miR-211-5p can increase SETBP1 level (six replicates per group, six independent experiments per group). (F) SETBP1 protein expression in MDA-MB-435 and MDA-MB-231 cells. ***P<0.001. Error bars represent the mean±s.e.m. A full colour version of this figure is available at the British Journal of Cancer journal online.
Expression of SETBP1 restored the inhibited proliferation and invasion function of miR-211-5p
We exogenously expressed recombinant SETBP1 lacking the 3′ UTR sequence (pcDNA3.1-SETBP1) in MDA-MB-231 cell. Western blotting showed that transfection of pcDNA3.1-SETBP1 and co-transfection of pcDNA3.1-SETBP1 and miR-211-5p increased the protein expression of SETBP1. Co-transfection of pcDNA3.1-SETBP1 and miR-211-5p abrogated the effects of miR-211-5p on SETBP1 expression (Figure 6A). Cells transfected with miR-211-5p showed significantly decreased levels of proliferation and invasion (Figure 6B and C). There are no significant differences of proliferation and invasion ability between cells transfected with pcDNA3.1-SETBP1 alone and cells co-transfected with pcDNA3.1-SETBP1 and miR-211-5p (Figure 6B and C). The results showed that expression of SETBP1 restored the inhibited proliferation and invasion function of miR-211-5p.
Expression of SETBP1 restored the inhibited proliferation and invasion function of miR-211-5p. (A) Cells were transfected with pcDNA3.1-SETBP1 (not including 3′UTR) and SETBP1 protein level was detected by western blot. GAPDH protein regarded as endogenous normaliser. (B) Cells were transfected with pcDNA3.1-SETBP1 (not including 3′ UTR) and miR-211-5p. Colony formation assays was performed (six replicates per group, three independent experiments per group). (C) Cells were transfected with pcDNA3.1-SETBP1 (not including 3′ UTR) and miR-211-5p (six replicates per group, three independent experiments per group). CCK-8 assays were performed. ***P<0.001. Error bars represent the mean±s.e.m. A full colour version of this figure is available at the British Journal of Cancer journal online.
SETBP1 mediated multiple biological actions of miR-211-5p in breast cancer cells
To determine whether SETBP1 acted as a critical mediator of miR-211-5p’s role in MDA-MB-231 cell, we performed targeted knockdown of SETBP1 expression using RNA interference-based short hairpin RNA. As illustrated in Figure 7A, western blot analysis showed a significant reduction of SETBP1 protein level in cells transfected with si-SETBP1 and cells transfected with miR-211-5p. As expected, the reduction of SETBP1 expression level led to a remarkable decrease in proliferation and invasion (Figure 7B and C). These results indicated that miR-211-5p regulated the proliferation and invasion of breast cancer cells, at least in part, by downregulating SETBP1. More importantly, we evaluated the prognostic value of miR-211-5p and SETBP1 expression in a public clinical database of TNBC, and found that high miR-211-5p expression was correlated with good prognosis, while high SETBP1 expression was associated with poor prognosis.
SETBP1 mediated multiple biological actions of miR-211-5p. (A) Cells were transfected with si-SETBP1 and miR-211-5p. SETBP1 protein level was detected by western blot. GAPDH protein regarded as endogenous normaliser. (B) Cells were transfected with si-SETBP1 and miR-211-5p. Colony formation assays were performed (six replicates per group, three independent experiments per group). (C) Cells were transfected with si-SETBP1 and miR-211-5p (six replicates per group, three independent experiments per group). CCK-8 assays were performed. ***P<0.001. (D) Kaplan–Meier representations of the probabilities of recurrence-free survival according to the expression levels of miR-211-5p and SETBP1. Error bars represent the mean±s.e.m. A full colour version of this figure is available at the British Journal of Cancer journal online.
Discussion
There is growing evidence that miRNAs play a crucial role in human cancer. MiRNA expression profiling has been utilised for classification of tumour stages and prognosis (van Schooneveld et al, 2012; Hofsli et al, 2013; Kanaan et al, 2013; Zheng et al, 2014). In the present study, we screened the miRNA expression pattern in TNBC tissues and serum using miRNA array and found that miR-211-5p was a TNBC-associated miRNA. Interestingly, miR-211-5p expression profile in TNBC tissues was identical to that in TNBC patients’ serum. Further studies demonstrated that miR-211-5p was significantly downregulated in TNBC. Importantly, miR-211-5p expression was significantly correlated with overall survival of TNBC. These findings implicate that miR-211-5p can be used as a prognostic biomarker for TNBC.
Recently, downregulation of miR-211-5p was found in cutaneous malignant melanomas and osteoarthritis (Kozubek et al, 2013; Prasadam et al, 2016). However, its biological role in breast cancer remains poorly understood. In the current study, we first demonstrated that miR-211-5p can inhibit TNBC cell proliferation, migration, invasion and metastasis both in vitro and in vivo, indicating the crucial role of miR-211-5p in TNBC.
It has been reported that abnormal expression of SETBP1 was associated with juvenile myelomonocytic leukaemia (Sakaguchi et al, 2013; Stieglitz et al, 2015). More importantly, the results of a genome-wide association study showed that SNPs in SETBP1 gene could contribute to the susceptibility for breast cancer (Michailidou et al, 2015). In these studies, SETBP1 was identified as an oncogene that contributed to breast cancer development. In our study, using a luciferase-based reporter assay, we demonstrated that miR-211-5p can bind to a sequence within the 3′-UTR of SETBP1. MiR-211-5p-mediated control of SETBP1 expression was further validated by complementary gain-of- and loss-of-function approaches. Importantly, overexpression of SETBP1 could effectively impede the ability of miR-211-5p to inhibit proliferation and metastasis. Our study provides solid evidence to support that miR-211-5p can inhibit proliferation and metastasis of TNBC by directly targeting SETBP1. A single protein, such as SETBP1, may not account for the multiple phenotypic effects (i.e., proliferation, apoptosis, migration and invasion). SETBP1 is a well characterised protein that is believed to inhibit PP2A phosphatase activity through SET stabilisation (Piazza et al, 2013). SETBP1 overexpression (due to lower level of miR-211-5p) protects SET from protease cleavage, increasing the amount of full-length SET protein that results in PP2A inhibition and therefore promotes proliferation and invasion. Importantly, PP2A dephosphorylates a broad range of proteins involved in multiple cellular process and dysregulation of PP2A has previously been implicated in TNBC (Tseng et al, 2012; Shu et al, 2016). A further study that specifically analyses the effect of miR-211-5p on SET and PP2A will be done in our following genetic studies.
Of note is the finding that overexpression of miR-211-5p can inhibit proliferation and metastasis of TNBC cells in mice, implicating miR-211-5p as a potential therapeutic target of TNBC. Intriguingly, our mechanistic and functional data permit us to better appreciate the functional role of SETBP1 in human breast cancer; its expression positively regulated cell proliferation, migration, invasion and metastasis. In addition, SETBP1, a direct functional target of miR-211-5p in TNBC, would be an attractive therapeutic target because it can be precisely targeted with a specific antibody. Targeting of deregulated miRNA is expected to be a well-tolerated therapy, because differentially expressed miRNAs are often disease-specific and do not play major roles under normal physiological conditions in adult tissues or in quiescent cells (Broderick and Zamore, 2011). In fact, miRNA knockout mice often do not display a specific phenotype, unless they are challenged with a stimulus causing cellular stress in target organs (Thum et al, 2008). Our main hypothesis derived from the present study is that increasing levels of miR-211-5p as a posttranscriptional regulator of SETBP1 could be a therapeutic strategy for TNBC. This attractive hypothesis needs to be further tested in animal models, and strategies to apply miRNA precursors or mature miRs in vivo have to be developed.
In conclusion, this study provides discovery and validation of TNBC-specific miRNA transcriptome profiles. We identified that miR-211-5p is a potent tumour suppressor in TNBC, and its growth inhibitory effects are, in part, mediated through its downstream target gene, SETBP1. Using cell culture and animal model, functional characterisation for the role of miR-211-5p reveals that loss of its expression is an event in TNBC progression.
Change history
27 June 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355.
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297.
Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V (2007) Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 109: 1721–1728.
Boyle P (2012) Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol 6: vi7–vi12.
Braunstein LZ, Taghian AG, Niemierko A, Salama L, Capuco A, Bellon JR, Wong JS, Punglia RS, MacDonald SM, Harris JR (2017) Breast-cancer subtype, age, and lymph node status as predictors of local recurrence following breast-conserving therapy. Breast Cancer Res Treat 161: 173–179.
Broderick JA, Zamore PD (2011) MicroRNA therapeutics. Gene Ther 18: 1104–1110.
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13: 4429–4434.
Dvinge H, Git A, Gräf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, Chin SF, Provenzano E, Turashvili G, Green A, Ellis I, Aparicio S, Caldas C (2013) The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497: 378–382.
Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjrn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S (2008) LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899.
Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple-negative breast cancer. N Engl J Med 363: 1938–1948.
Hofsli E, Sjursen W, Prestvik WS, Johansen J, Rye M (2013) Identification of serum microRNA profiles in colon cancer. Br J Cancer 108: 1712–1719.
Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S (2013) A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg 258: 400–408.
Kim BG, Kang S, Han HH, Lee JH, Kim JE, Lee SH, Cho NH (2016) Transcriptome-wide analysis of compression-induced microRNA expression alteration in breast cancer for mining therapeutic targets. Oncotarget 7: 27468–27478.
Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017.
Kozubek J, Ma Z, Fleming E, Duggan T, Wu R, Shin DG, Dadras SS (2013) In-depth characterization of microRNA transcriptome in melanoma. PLoS ONE 8: e72699.
Kwilas AR, Ardiani A, Gameiro SR, Richards J, Hall AB, Hodge JW (2016) Androgen deprivation therapy sensitizes triple negative breast cancer cells to immune-mediated lysis through androgen receptor independent modulation of osteoprotegerin. Oncotarget 7: 23498–23511.
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumor invasion and metastasis initiated by microRNA-10b in breast cancer. Nature (2007); 449: 682–688.
Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah M, Perkins BJ, Czene K, Eriksson M, Darabi H, Brand JS, Bojesen SE, Nordestgaard BG, Flyger H, Nielsen SF, Rahman N, Turnbull C, BOCS, Fletcher O, Peto J, Gibson L, dos-Santos-Silva I, Chang-Claude J, Flesch-Janys D, Rudolph A, Eilber U, Behrens S, Nevanlinna H, Muranen TA, Aittomki K, Blomqvist C, Khan S, Aaltonen K, Ahsan H, Kibriya MG, Whittemore AS, John EM, Malone KE, Gammon MD, Santella RM, Ursin G, Makalic E, Schmidt DF, Casey G, Hunter DJ, Gapstur SM, Gaudet MM, Diver WR (2015) Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet 47: 373–380.
Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, Antolini L, Mologni L, Donadoni C, Papaemmanuil E, Schnittger S, Kim DW, Boultwood J, Rossi F, Gaipa G, De Martini GP, di Celle PF, Jang HG, Fantin V, Bignell GR, Magistroni V, Haferlach T, Pogliani EM, Campbell PJ, Chase AJ, Tapper WJ, Cross NC, Gambacorti-Passerini C (2013) Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet 45: 18–24.
Prasadam I, Batra J, Perry S, Gu W, Crawford R, Xiao Y (2016) Systematic identification, characterization and target gene analysis of microRNAs involved in osteoarthritis subchondral bone pathogenesis. Calcif Tissue Int 99: 43–55.
Prat A, Lluch A, Albanell J, Barry WT, Fan C, Chacón JI, Parker JS, Calvo L, Plazaola A, Arcusa A, Seguí-Palmer MA, Burgues O, Ribelles N, Rodriguez-Lescure A, Guerrero A, Ruiz-Borrego M, Munarriz B, López JA, Adamo B, Cheang MC, Li Y, Hu Z, Gulley ML, Vidal MJ, Pitcher BN, Liu MC, Citron ML, Ellis MJ, Mardis E, Vickery T, Hudis CA, Winer EP, Carey LA, Caballero R, Carrasco E, Martín M, Perou CM, Alba E (2014) Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br J Cancer 111: 1532–1541.
Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, Kon A, Sanada M, Chiba K, Tanaka H, Makishima H, Wang X, Xu Y, Doisaki S, Hama A, Nakanishi K, Takahashi Y, Yoshida N, Maciejewski JP, Miyano S, Ogawa S, Kojima S (2013) Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet 45: 937–941.
Shin VY, Siu JM, Cheuk I, Ng EK, Kwong A (2015) Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br J Cancer 112: 1751–1759.
Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Peluffo G, Brown J, D'Santos C, Krop IE, Dillon D, McKeown M, Ott C, Qi J (2016) Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529: 413–417.
Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Peluffo G, Brown J, D'Santos C, Krop IE, Dillon D, McKeown M, Ott C, Qi J, Ni M, Rao PK, Duarte M, Wu SY, Chiang CM, Anders L, Young RA, Winer EP, Letai A, Barry WT, Carroll JS, Long HW, Brown M, Liu XS, Meyer CA, Bradner JE, Polyak K (2016) Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529: 413–417.
Stieglitz E, Troup CB, Gelston LC, Haliburton J, Chow ED, Yu KB (2015) Subclonal mutations in SETBP1 confer a poor prognosis in juvenile myelomonocytic leukemia. Blood 125: 516–524.
Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456: 980–984.
Tseng LM, Liu CY, Chang KC, Chu PY, Shiau CW, Chen KF (2012) CIP2A is a target of bortezomib in human triple negative breast cancer cells. Breast Cancer Res 14: R68.
van Roozendaal LM, Smit LH, Duijsens GH, de Vries B, Siesling S, Lobbes MB (2016) Risk of regional recurrence in triple-negative breast cancer patients: a Dutch cohort study. Breast Cancer Res Treat 156: 465–472.
van Schooneveld E, Wouters MC, Van der Auwera I, Peeters DJ, Wildiers H, Van Dam PA, Vergote I (2012) Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res 14: R34.
Verghese ET, Drury R, Green CA, Holliday DL, Lu X, Nash C, Speirs V, Thorne JL, Thygesen HH, Zougman A, Hull MA, Hanby AM, Hughes TA (2013) MiR-26b is down-regulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion. J Pathol 231: 388–399.
Woolston C (2015) Breast cancer. Nature 527: S101.
Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham D, Glennie MJ, Kelland LR, Robinson V, Stratford IJ, Tozer GM, Watson S, Wedge SR, Eccles SA Committee of the National Cancer Research Institute (2010) Guidelines for the welfare and use of animals in cancer research. Br J Cancer 102: 1555–1577.
Yaffe MJ, Jong RA (2016) Adjunctive ultrasonography in breast cancer screening. Lancet 387: 313–314.
Yates LA, Norbury CJ, Gilbert RJ (2013) The long and short of microRNA. Cell 153: 516–519.
Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, Li J, Wang C (2014) Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer 111: 1985–1992.
Acknowledgements
We thank all donors enroled in the present study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Chen, Ll., Zhang, Zj., Yi, Zb. et al. MicroRNA-211-5p suppresses tumour cell proliferation, invasion, migration and metastasis in triple-negative breast cancer by directly targeting SETBP1. Br J Cancer 117, 78–88 (2017). https://doi.org/10.1038/bjc.2017.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.150
Keywords
This article is cited by
-
Meta-analysis of integrated ChIP-seq and transcriptome data revealed genomic regions affected by estrogen receptor alpha in breast cancer
BMC Medical Genomics (2023)
-
miRNA-211 maintains metabolic homeostasis in medulloblastoma through its target gene long-chain acyl-CoA synthetase 4
Acta Neuropathologica Communications (2023)
-
Identification of a chromatin regulator signature and potential candidate drugs for bladder cancer
Hereditas (2022)
-
The functional significance and cross-talk of non-coding RNAs in triple negative and quadruple negative breast cancer
Molecular Biology Reports (2022)
-
A Proposed TUSC7/miR-211/Nurr1 ceRNET Might Potentially be Disturbed by a cer-SNP rs2615499 in Breast Cancer
Biochemical Genetics (2022)