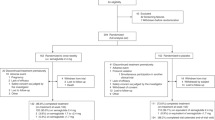
Abstract
Metabolic syndrome after transplantation is a major concern following solid organ transplantation (SOT). The CREB-regulated transcription co-activator 2 (CRTC2) regulates glucose metabolism. The effect of CRTC2 polymorphisms on new-onset diabetes after transplantation (NODAT) was investigated in a discovery sample of SOT recipients (n1=197). Positive results were tested for replication in two samples from the Swiss Transplant Cohort Study (STCS, n2=1294 and n3=759). Obesity and other metabolic traits were also tested. Associations with metabolic traits in population-based samples (n4=46’186, n5=123’865, n6>100,000) were finally analyzed. In the discovery sample, CRTC2 rs8450-AA genotype was associated with NODAT, fasting blood glucose and body mass index (Pcorrected<0.05). CRTC2 rs8450-AA genotype was associated with NODAT in the second STCS replication sample (odd ratio (OR)=2.01, P=0.04). In the combined STCS replication samples, the effect of rs8450-AA genotype on NODAT was observed in patients having received SOT from a deceased donor and treated with tacrolimus (n=395, OR=2.08, P=0.02) and in non-kidney transplant recipients (OR=2.09, P=0.02). Moreover, rs8450-AA genotype was associated with overweight or obesity (n=1215, OR=1.56, P=0.02), new-onset hyperlipidemia (n=1007, OR=1.76, P=0.007), and lower high-density lipoprotein-cholesterol (n=1214, β=-0.08, P=0.001). In the population-based samples, a proxy of rs8450G>A was significantly associated with several metabolic abnormalities. CRTC2 rs8450G>A appears to have an important role in the high prevalence of metabolic traits observed in patients with SOT. A weak association with metabolic traits was also observed in the population-based samples.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Mayer AD, Dmitrewski J, Squifflet JP, Besse T, Grabensee B, Klein B et al. Multicenter randomized trial comparing tacrolimus (FK506) and cyclosporine in the prevention of renal allograft rejection: a report of the European Tacrolimus Multicenter Renal Study Group. Transplantation 1997; 64: 436–443.
Hariharan S, Johnson CP, Bresnahan BA, Taranto SE, McIntosh MJ, Stablein D . Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med 2000; 342: 605–612.
Ojo AO, Hanson JA, Wolfe RA, Leichtman AB, Agodoa LY, Port FK . Long-term survival in renal transplant recipients with graft function. Kidney Int 2000; 57: 307–313.
Collins AJ, Foley RN, Herzog C, Chavers B, Gilbertson D, Ishani A et al. United States Renal Data System 2008 Annual Data Report. Am J Kidney Dis 2009; 53: S1–S374.
Johnston SD, Morris JK, Cramb R, Gunson BK, Neuberger J . Cardiovascular morbidity and mortality after orthotopic liver transplantation. Transplantation 2002; 73: 901–906.
Vogt DP, Henderson JM, Carey WD, Barnes D . The long-term survival and causes of death in patients who survive at least 1 year after liver transplantation. Surgery 2002; 132: 775–780.
Opelz G, Dohler B . Collaborative Transplant S. Influence of immunosuppressive regimens on graft survival and secondary outcomes after kidney transplantation. Transplantation 2009; 87: 795–802.
First MR, Dhadda S, Croy R, Holman J, Fitzsimmons WE . New-onset diabetes after transplantation (NODAT): an evaluation of definitions in clinical trials. Transplantation 2013; 96: 58–64.
Kasiske BL, Snyder JJ, Gilbertson D, Matas AJ . Diabetes mellitus after kidney transplantation in the United States. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons 2003; 3: 178–185.
Israni AK, Snyder JJ, Skeans MA, Kasiske BL, Investigators P. Clinical diagnosis of metabolic syndrome: predicting new-onset diabetes, coronary heart disease, and allograft failure late after kidney transplant. Transplant international: official journal of the European Society for Organ Transplantation 2012; 25: 748–757.
Pham PT, Pham PM, Pham SV, Pham PA, Pham PC . New onset diabetes after transplantation (NODAT): an overview. Diabetes, metabolic syndrome and obesity: targets and therapy 2011; 4: 175–186.
Billings LK, Florez JC . The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci 2010; 1212: 59–77.
Torres JM, Cox NJ, Philipson LH . Genome wide association studies for diabetes: perspective on results and challenges. Pediatr Diabetes 2013; 14: 90–96.
McCaughan JA, McKnight AJ, Maxwell AP. Genetics of new-onset diabetes after transplantation. J Am Soc Nephrol 2014; 25: 1037–1049.
Chand S, Shabir S, Chan W, McCaughan JA, McKnight AJ, Maxwell AP et al. beta cell glucotoxic-associated single nucleotide polymorphisms in impaired glucose tolerance and new-onset diabetes after transplantation. Transplantation 2014; 98: e19–e20.
Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 2005; 437: 1109–1111.
Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB et al. TORCs: transducers of regulated CREB activity. Mol Cell 2003; 12: 413–423.
Choong E, Quteineh L, Cardinaux JR, Gholam-Rezaee M, Vandenberghe F, Dobrinas M et al. Influence of CRTC1 polymorphisms on body mass index and fat mass in psychiatric patients and the general adult population. JAMA Psychiatry 2013; 70: 1011–1019.
Canettieri G, Koo SH, Berdeaux R, Heredia J, Hedrick S, Zhang X et al. Dual role of the coactivator TORC2 in modulating hepatic glucose output and insulin signaling. Cell Metab 2005; 2: 331–338.
Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 2004; 119: 61–74.
Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X et al. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol 2004; 14: 2156–2161.
Eberhard CE, Fu A, Reeks C, Screaton RA . CRTC2 is required for beta-cell function and proliferation. Endocrinology 2013; 154: 2308–2317.
Keshavarz P, Inoue H, Nakamura N, Yoshikawa T, Tanahashi T, Itakura M . Single nucleotide polymorphisms in genes encoding LKB1 (STK11), TORC2 (CRTC2) and AMPK alpha2-subunit (PRKAA2) and risk of type 2 diabetes. Mol Genet Metab 2008; 93: 200–209.
He Y, Li Y, Qiu Z, Zhou B, Shi S, Zhang K et al. Identification and validation of PROM1 and CRTC2 mutations in lung cancer patients. Mol Cancer 2014; 13: 19.
Crettol S, Venetz JP, Fontana M, Aubert JD, Pascual M, Eap CB . CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients. Ther Drug Monit 2008; 30: 689–699.
Crettol S, Venetz JP, Fontana M, Aubert JD, Ansermot N, Fathi M et al. Influence of ABCB1 genetic polymorphisms on cyclosporine intracellular concentration in transplant recipients. Pharmacogenet Genomics 2008; 18: 307–315.
Davidson J, Wilkinson A, Dantal J, Dotta F, Haller H, Hernandez D et al. New-onset diabetes after transplantation: 2003 International consensus guidelines. Proceedings of an international expert panel meeting. Barcelona, Spain, 19 February 2003. Transplantation 2003; 75: SS3–S24.
Koller MT, van Delden C, Muller NJ, Baumann P, Lovis C, Marti HP et al. Design and methodology of the Swiss Transplant Cohort Study (STCS): a comprehensive prospective nationwide long-term follow-up cohort. Eur J Epidemiol 2013; 28: 347–355.
Manuel O, Kralidis G, Mueller NJ, Hirsch HH, Garzoni C, van Delden C et al. Impact of antiviral preventive strategies on the incidence and outcomes of cytomegalovirus disease in solid organ transplant recipients. Am J Transplant 2013; 13: 2402–2410.
Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009; 41: 77–81.
Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010; 467: 832–838.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948.
Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet 2010; 42: 949–960.
Genome Wide Associations Scans for Total Cholesterol, HDL-C, LDL-C and triglycerides. Available at: http://www.sph.umich.edu/csg/abecasis/public/lipids2010/ [accessed on 14 January 2014].
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010; 466: 707–713.
He C, Holme J, Anthony J . SNP genotyping: the KASP assay. Methods Mol Biol 2014; 1145: 75–86.
Altarejos JY, Goebel N, Conkright MD, Inoue H, Xie J, Arias CM et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat Med 2008; 14: 1112–1117.
Song Y, Altarejos J, Goodarzi MO, Inoue H, Guo X, Berdeaux R et al. CRTC3 links catecholamine signalling to energy balance. Nature 2010; 468: 933–939.
Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI . SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008; 24: 2938–2939.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797.
Rodrigo E, Fernandez-Fresnedo G, Valero R, Ruiz JC, Pinera C, Palomar R et al. New-onset diabetes after kidney transplantation: risk factors. J Am Soc Nephrol 2006; 17: S291–S295.
Numakura K, Satoh S, Tsuchiya N, Horikawa Y, Inoue T, Kakinuma H et al. Clinical and genetic risk factors for posttransplant diabetes mellitus in adult renal transplant recipients treated with tacrolimus. Transplantation 2005; 80: 1419–1424.
Gourishankar S, Jhangri GS, Tonelli M, Wales LH, Cockfield SM . Development of diabetes mellitus following kidney transplantation: a Canadian experience. Am J Transplant 2004; 4: 1876–1882.
Sulanc E, Lane JT, Puumala SE, Groggel GC, Wrenshall LE, Stevens RB . New-onset diabetes after kidney transplantation: an application of 2003 International Guidelines. Transplantation 2005; 80: 945–952.
Maes BD, Kuypers D, Messiaen T, Evenepoel P, Mathieu C, Coosemans W et al. Post transplantation diabetes mellitus in FK-506-treated renal transplant recipients: analysis of incidence and risk factors. Transplantation 2001; 72: 1655–1661.
Acknowledgements
Data on glycaemic traits have been contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. CBE takes full responsibility for the work as a whole, including the study design, access to data, and the decision to submit and publish the manuscript. This work has been funded in part by the Swiss National Science Foundation (CBE: 324730_144064). LQ and CBE received research support from the Roche Organ Transplantation Research Foundation (#152358701) and the STCS in the past 3 years. ZK was funded by the Swiss National Science Foundation (31003A-143914) and the Leenaards Foundation. This study has been conducted in the framework of the STCS, supported by the Swiss National Science Foundation and the Swiss University Hospitals (G15) and transplant centers.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
CBE received honoraria for conferences or teaching CME courses from Advisis, Astra Zeneca, Lundbeck, MSD, Sandoz, Servier and Vifor-Pharma in the past 3 years. He received an unrestricted educational grant from Takeda in the past 3 years. JFD Advisory committees: Bayer, BMS, Gilead Science, Janssen Cilag, Jennerex, Merck, Novartis, Roche. Speaking and teaching: Bayer, Boehringer-Ingelheim, Novartis, Roche. SC received honoraria for teaching CME courses from Astra Zeneca and Lundbeck. The remaining authors have no conflict of interest.
Additional information
Swiss Transplant Cohort Study Rita Achermann, John-David Aubert, Philippe Baumann, Guido Beldi, Christian Benden, Christoph Berger, Isabelle Binet, Pierre-Yves Bochud, Elsa Boely (Head of local data management), Heiner Bucher, Leo Bühler, Thierry Carell, Emmanuelle Catana, Yves Chalandon, Sabina de Geest, Olivier de Rougemont, Michael Dickenmann, Michel Duchosal, Thomas Fehr, Sylvie Ferrari-Lacraz, Christian Garzoni, Yvan Gasche, Paola Gasche Soccal, Emiliano Giostra, Déla Golshayan, Daniel Good, Karine Hadaya, Christoph Hess, Sven Hillinger, Hans Hirsch, Günther Hofbauer, Uyen Huynh-Do, Franz Immer, Richard Klaghofer, Michael Koller (Head of the data center), Thomas Kuntzen, Bettina Laesser, Roger Lehmann, Christian Lovis, Oriol Manuel, Hans-Peter Marti, Pierre Yves Martin, Pascal Meylan (Head, Biological samples management group), Paul Mohacsi, Isabelle Morard, Philippe Morel, Ulrike Mueller, Nicolas Mueller (Chairman Scientific Committee), Helen Mueller-McKenna, Thomas Müller, Beat Müllhaupt, David Nadal, Gayathri Nair, Manuel Pascual (Executive office), Jakob Passweg, Chantal Piot Ziegler, Juliane Rick, Eddy Roosnek, Anne Rosselet, Silvia Rothlin, Frank Ruschitzka, Urs Schanz, Stefan Schaub, Christian Seiler, Nasser Semmo, Susanne Stampf, Jürg Steiger (Head, Executive Office), Christian Toso, Dimitri Tsinalis, Christian Van Delden (Executive office), Jean-Pierre Venetz, Jean Villard, Madeleine Wick (STCS coordinator), Markus Wilhelm, Patrick Yerly.
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Quteineh, L., Bochud, PY., Golshayan, D. et al. CRTC2 polymorphism as a risk factor for the incidence of metabolic syndrome in patients with solid organ transplantation. Pharmacogenomics J 17, 69–75 (2017). https://doi.org/10.1038/tpj.2015.82
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2015.82
This article is cited by
-
Donor and recipient polygenic risk scores influence the risk of post-transplant diabetes
Nature Medicine (2022)
-
Sam68 promotes hepatic gluconeogenesis via CRTC2
Nature Communications (2021)
-
Genetic and clinic predictors of new onset diabetes mellitus after transplantation
The Pharmacogenomics Journal (2019)