Abstract
Eye movement deviations, particularly deficits of initial sensorimotor processing and sustained pursuit maintenance, and antisaccade inhibition errors, are established intermediate phenotypes for psychotic disorders. We here studied eye movement measures of 849 participants from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study (schizophrenia N=230, schizoaffective disorder N=155, psychotic bipolar disorder N=206 and healthy controls N=258) as quantitative phenotypes in relation to genetic data, while controlling for genetically derived ancestry measures, age and sex. A mixed-modeling genome-wide association studies approach was used including ~4.4 million genotypes (PsychChip and 1000 Genomes imputation). Across participants, sensorimotor processing at pursuit initiation was significantly associated with a single nucleotide polymorphism in IPO8 (12p11.21, P=8 × 10−11), whereas suggestive associations with sustained pursuit maintenance were identified with SNPs in SH3GL2 (9p22.2, P=3 × 10−8). In participants of predominantly African ancestry, sensorimotor processing was also significantly associated with SNPs in PCDH12 (5q31.3, P=1.6 × 10−10), and suggestive associations were observed with NRSN1 (6p22.3, P=5.4 × 10−8) and LMO7 (13q22.2, P=7.3x10−8), whereas antisaccade error rate was significantly associated with a non-coding region at chromosome 7 (P=6.5 × 10−9). Exploratory pathway analyses revealed associations with nervous system development and function for 40 top genes with sensorimotor processing and pursuit maintenance (P=4.9 × 10−2–9.8 × 10−4). Our findings suggest novel patterns of genetic variation relevant for brain systems subserving eye movement control known to be impaired in psychotic disorders. They include genes involved in nuclear trafficking and gene silencing (IPO8), fast axonal guidance and synaptic specificity (PCDH12), transduction of nerve signals (NRSN1), retinal degeneration (LMO7), synaptic glutamate release (SH3GL2), and broader nervous system development and function.
Similar content being viewed by others
Introduction
Deviations of eye movement control are established neurophysiological intermediate phenotypes for psychotic disorders that may be useful for advancing gene discovery in psychiatry.1 Impairments are seen in a reduced ability to accurately track slowly moving objects with the eyes2 and to voluntarily suppress a reflexive saccade to a peripheral target on antisaccade tasks.3, 4 Consistent with multiple lines of evidence indicating shared neurobiological alterations and genetic vulnerability across schizophrenia spectrum and psychotic bipolar disorders,5, 6, 7, 8, 9 comparable eye movement deficits have been demonstrated across these groups in first-episode and chronically ill patients, and in their relatives indicating disturbances in brain systems subserving pursuit initiation and maintenance, and inhibitory control.2, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 We recently reported both smooth pursuit impairments and antisaccade inhibition errors in a large cohort of clinically stabilized psychotic disorder cases and their relatives as part of the Bipolar and Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Consortium Study.28, 29, 30 We found that the initiation of a pursuit movement, which depends on rapid sensorimotor processing, was disturbed in probands and their relatives, while pursuit maintenance, dependent on cognitive predictions of target motion and the most widely used phenotype in prior genetic studies, was mostly impaired in probands.29 Impaired antisaccade task performance was identified in probands and their relatives, reflecting decreased inhibitory behavioral control.28 How these intermediate phenotypes are related to genetic variation across the genome has to date not been comprehensively studied.
The first genetic studies of eye movement abnormalities in psychotic disorders reported linkage between pursuit maintenance ability and microsatellite markers on the short arm of chromosome 6 (6p21-23).31, 32 Subsequent genetic studies using eye movement phenotypes have predominantly focused on single nucleotide polymorphisms (SNPs) in candidate genes for schizophrenia disease risk, for example, catechol-O-methyltransferase and neuregulin-1.33, 34, 35, 36, 37, 38, 39 Lencer et al.40 reported an association of pursuit-initiation impairments in first-episode psychosis patients with a dopamine D2 receptor gene (DRD2), whereas pursuit maintenance was associated to candidate SNPs in metabotropic glutamate receptor 3 protein (GRM3). This finding supports a model of different genes being potentially significant for different aspects of eye movement control.
Despite these initial reports, confirmation and larger scale genome-wide association studies (GWAS) in patients with psychosis are lacking. We report herein a GWAS evaluating genetic associations with three eye movement phenotypes representing (1) initial sensorimotor processing (pursuit acceleration), (2) sustained pursuit (maintenance gain) and (3) voluntary inhibitory control (antisaccade error rate) in probands with psychosis and controls from the B-SNIP sample, with additional exploratory pathway analyses to identify biological networks implicated by top findings.
Materials and methods
Participants
Smooth pursuit and antisaccade measures were assessed in 849 participants (schizophrenia N=230, schizoaffective disorder N=155, psychotic bipolar disorder N=206 and healthy controls N=258) of the B-SNIP consortium for whom DNA and genotyping information were available. In depth descriptions of the overall B-SNIP study design, inclusion and exclusion criteria, clinical ratings and eye movement assessments have been previously described.28, 29, 30 Diagnoses were made by a consensus process using all available clinical information including the Structured Clinical Interview for DSM IV (SCID)41 with collateral information from family members when available. Probands were clinically stable and receiving consistent psychopharmacological treatment for at least 1 month (Table 1; Supplementary Table 1).42, 43, 44, 45, 46, 47
Inclusion criteria for all subjects were (1) age 15–65; (2) WRAT reading score ⩾65;42 (3) no history of neurologic or systemic disease; (4) minimum of 20/40 visual acuity (with or without correction) and (5) no history of substance abuse within the last month or substance dependence within the last three months according to SCID, and negative urine toxicology (MP On-Site 11: One Step Onsite, ref: 60B02-MPB) on assessment day. Inclusion criteria for control subjects additionally included: (1) no personal or family history (first-degree) of psychotic or bipolar disorder; (2) no history of recurrent depression; and (3) no history of psychosis spectrum personality traits defined as meeting full or within one criteria of a cluster A (psychosis spectrum) Axis-II diagnosis. The study was approved by institutional review boards at each study site and written informed consent was obtained prior to study participation.
Eye movement analyses
The eye movement measures (Table 1) utilized as primary outcome measures in genetic analyses included: (1) initial pursuit acceleration (measure of rapid sensorimotor processing during the first 100ms of pursuit assessed by foveo-petal step-ramp stimuli (18.7°/s);29 (2) pursuit maintenance gain (accuracy of matching eye to target velocity during sustained pursuit) using a triangular wave task (18.7°/s);29 and (3) antisaccade error rate defined as the percentage of trials with failed response inhibition from an overlap task,28 (Supplementary Material Methods). Eye movements were acquired with a video-based eye tracker in a darkened room (Eyelink II, SR Research, Ottawa, ON, Canada, sampling rate 500 Hz) with the same testing conditions and hardware used at all B-SNIP sites. Each eye movement measure was standardized using a normative regression approach, transforming data to z-scores including age, race and sex as covariates. This was done to remove variance in data related to demographic parameters from all groups in a similar way, and to facilitate comparison of the magnitude of effects across the different groups and pursuit measures. Our previous analyses with the B-SNIP study sample did not identify significant effects of antipsychotic dosing, anticholinergic loading or other medication effects on eye movement measures in these stably treated patients.28, 29 Furthermore, eye movement measures were shown to be relatively independent from general cognitive deficits indicated by BACS scores.28, 29
Genotyping and imputation
Genomic DNA from participants was isolated from whole blood using standard protocols and genotyped by the Broad Institute using the Illumina Infinium PsychChip array. Quality control (QC) procedures were conducted with PLINK v1.948 following standardized protocols.49 Genetic markers deviating from Hardy–Weinberg Equilibrium (P<10E−6), genotype-inferred sex differing from reported sex, or having call rates <98% were excluded from analyses. We included SNPs that had minor allele frequencies (MAF) ⩾0.01 in case or control groups. Cryptic relatedness was checked with PREST-plus.50 Samples showing a second degree relationship or greater were excluded resulting in 849 participants available for GWAS.
SNPs passing quality control procedures were used for imputation using HAPI-UR for pre-phasing51 and IMPUTE2 for imputation52 using the 1000 Genomes phase 1 data as a reference panel.53 Poorly imputed SNPs were filtered with the resulting imputed SNPs merged back in with the directly genotyped SNPs from the PsychChip for a total of 4 404 269 SNPs passing filtering criteria used for the analyses described herein.
Genetic ancestry assessments were completed with multi-dimensional scaling (MDS) plots relative to 1000 Genomes Project populations. Race stratified analyses represented a division of the two predominating ancestry components. Analyses of both stratified and whole group analyses utilized the first five principle components of ancestry analyses as covariates.
Genome-wide association analyses approach
We used a mixed-modeling approach as implemented in the Efficient Mixed-Model Association eXpedited (EMMAX)-software package,54 which uses an identity by state (IBS) relationship matrix, and the first five eigenvectors from principle components analysis (PCA) included as covariates to reliably account for mixed ethnicity populations. Standardized eye movement measures (see above) were modeled as quantitative trait phenotypes in relation to genetic data. Probands and controls were grouped together for primary analyses with all ancestry groups combined. For secondary analyses, the sample was stratified by the top two genetically derived ancestry groups with follow-up studies in the proband only sample. The rationale for grouping cases and controls together in primary analyses was to take advantage of the wider range of phenotypic variance for the examination of genetic contributions to eye movement control.
To account for multiple testing using imputed data, the genome-wide significance threshold was set at 1 x E−08, which is more conservative than the commonly used GWAS significance threshold of 5 x E−08.55 False discovery rate (FDR) q-statistics further adjusting for multiple analyses of phenotypes and race groups were calculated. FDR q-values for GWAS significant findings remained <0.05 with the exception of rs2010148567 in relation to antisaccade response inhibition in African ancestry (AA), where q=0.09, all collectively indicating low type I error. We define ‘suggestive associations’ as P-values exceeding 5 x E−7 but not meeting 1 x E−8 GWAS significance. The closest gene was assigned to each SNP using BEDTools closest and RefSeq gene annotations from hg19.56
Exploratory pathway analyses
We used Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA, USA) to conduct exploratory analyses (using the Core Analysis feature) of genes affiliated (within 15 kb) with the top 200 SNP associations identified through GWAS analyses. Associations in all participants were examined separately for each eye movement measure by merging the top 200 associated SNPs from the two primary ancestry group analyses. The expression quantitative trait loci analyses of top SNPs associated with clinical phenotypes were performed using the Genotype-Tissue Expression GTEx Portal (www.gtexportal.org/home) and the United Kingdom Brain Expression Consortium (UKBEC, www.braineac.org).
Results
Initial sensorimotor processing
GWAS of initial pursuit acceleration in all participants
Across participants, the most robust genome-wide significant association was identified with an isolated SNP in the Importin 8 gene (IPO8) at chromosome 12p11.21 (Table 2). In addition, a number of SNPs in a non-coding RNA gene at chromosome 2p12, and in an intergenic region near the mitogen-activated protein kinase MAP3K1 gene at chromosome 5q11.2 showed patterns of suggestive association.
GWAS of initial pursuit acceleration stratified by ancestry
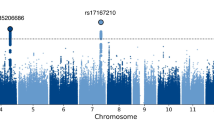
GWAS in participants of predominantly AA (N=300) revealed the aforementioned genome-wide significant association with IPO8, as well as an additional genome-wide significant association with SNPs in the protocadherin 12 (PCDH12) gene at chromosome 5q31.3 (Figure 1a; Table 2). Other polymorphisms with suggestive associations included SNPs ~300 kb upstream of the Neurensin 1 gene (NRSN1) in an intergenic region at chromosome 6p22.3 and a SNP in the LIM domain only protein 7 gene (LMO7) at chromosome 13q22.2.
Manhattan plots from genome-wide association studies (GWAS) stratified for participants of predominantly African ancestry (N=300, left side) and participants of predominantly Caucasian ancestry (N=549, right side). Results for the three eye movement measures used as phenotypes in GWAS are depicted: (a) initial pursuit acceleration, (b) pursuit maintenance gain and (c) antisaccade error rate. For more details see Table 2.
In participants of predominantly Caucasian ancestry (CA, N=549), initial pursuit acceleration was significantly associated with a SNP representing a missense mutation in CYB5R3 at chromosome 22q13.2 coding for membrane bound cytochrome B5 reductase 3. In addition, the aforementioned SNPs in a non-coding RNA gene at chromosome 2p12 showed suggestive associations.
GWAS of initial pursuit acceleration in probands only
Follow-up analyses in probands across ancestries identified the genome-wide significant association with the SNP in IPO8 that was seen in the whole-study sample, and additionally suggestive association in LMO7. Similarly, follow-up analyses in AA probands revealed the genome-wide significant associations with SNPs in IPO8, PCDH12 and LMO7 (Table 3). In the sub-sample of CA probands, no genome-wide significant association was observed with initial pursuit acceleration.
Sustained pursuit maintenance
GWAS of maintenance gain in all participants
No associations were identified which exceeded our pre-defined threshold for genome-wide significance of 1 × E−8.55 Suggestive associations with pursuit maintenance gain across all participants were identified with a number of SNPs in or around the src Homology-3 (SH3) domain gene (SH3GL2) at chromosome 9p22.2 (Table 2).
GWAS of pursuit maintenance gain stratified by ancestry
Suggestive associations with SH3GL2 were also observed in CA participants only (Table 2; Figure 1b). In AA participants, we identified further suggestive associations with pursuit maintenance ability. These included SNPs in TMPRSS5 at chromosome 11q23.1 encoding a transmembrane serine protease, and in POP7 at chromosome 7q22.1, which is a protein-coding gene related to gene expression and RNA transport. Very close to this region on chromosome 7, we additionally identified a SNP in GGYF1, which encodes a protein believed to act cooperatively with growth factor receptor-bound protein10 (GRB10) to regulate tyrosine kinase receptor signaling.
Follow-up analyses in the proband subsample (Table 3) showed suggestive associations of pursuit maintenance gain with SNPs in POP7, GGYF1 and TMPRSS5 in the subsample of AA probands but not in CA probands.
GWAS of antisaccade error rates
GWAS with antisaccade error rate in AA participants identified one GWAS significant SNP and 25 suggestive SNPs in an intergenic region at chromosome 7 (Table 2; Figure 1c). However, no further associations with error rate were observed in either the whole sample or proband subsamples considered separately.
More details on top 200 SNPs identified in primary and secondary GWAS are given in Supplementary Table 2.
Exploratory pathway analyses
The top 200 SNP associations identified in race stratified GWAS analyses represented 89 distinct genes for initial pursuit acceleration and 103 distinct genes for pursuit maintenance gain (Supplementary Table 3). A top physiological system category identified for both pursuit phenotypes was nervous system development and function, which was represented by ~19% (N=17) of the top genes associated with initial pursuit acceleration and ~22% (N=23) of the top genes associated with pursuit maintenance gain (enrichment P-value range 4.9−10−2–9.8 × 10−4). Noting the similarities in pathway relationships identified, the genes comprising these lists were merged (N=189 unique genes; N=40 genes relevant to the nervous system) and visualized with neural network mapping that highlights the nervous system development and synaptic functioning (Figure 2). This revealed functions including ‘formation of the eye’, ‘eyelid reflex’ and ‘electrophysiology of the eye’; ‘excitatory postsynaptic action potential’; as well as ‘neurological signs’, ‘movement disorders’ and ‘neurodegeneration’. Nineteen of these genes have previously shown evidence for a relationship to psychotic disorders.
Summary of Ingenuity Pathway Analysis (IPA) using genes encoding the top 200 SNPs associated with initial pursuit acceleration and pursuit maintenance within the study population (N=849). The functional category nervous system development and function was identified as one of the top five physiological systems represented by these genes. Genes listed from the top have the greatest number of connections to functional categories within the nervous system development category listed in the lower panel. Genes that have shown evidence for a relationship to psychotic disorders are highlighted in red. SNP, single-nucleotide polymorphism.
Discussion
In this GWAS, we focused on eye movement measures indexing different neurophysiological aspects of eye movement control known to be impaired in psychotic disorders. We identified novel genome-wide significant findings that may promote understanding of psychosis risk and pathophysiology. First, the most significant associations were found for initial sensorimotor processing with IPO8 at 12p11.21, PCDH12 at 5q31.3, CYB5R3 at 22q13.2 and LMO7 at 13q22.2. These associations were predominantly observed in variants with lower minor frequencies, mostly in AA participants. Second, suggestive associations with sustained pursuit maintenance were observed with protein coding SNPs in and around SH3GL2 at 9p22.2. Third, significant genome-wide association of behavioral response inhibition was observed with a non-coding region at chromosome 7 in AA participants. All genes for which we found significant associations with referring SNPs are expressed in the brain (www.gtexportal.org/home; www.braineac.org). Those variants exceeding our pre-defined GWAS significance threshold also had low FDR statistics after accounting for multiple comparisons. Finally, exploratory pathway analyses of top associated SNPs identified commonalities between genes related to smooth pursuit measures, which consisted of loci previously associated with brain development, neurophysiology, ocular physiology and schizophrenia risk. These findings provide important new genetic information about what has long been one of the most promising familial phenotypes associated with psychotic disorders.1, 2, 57 This said, our findings extend previous reports from large-scale genetic studies showing considerable overlap between schizophrenia spectrum and bipolar disorders.6, 7, 8, 9
Genetic alterations related to initial sensorimotor processing
The IPO8 gene, in which we found a missense mutation significantly associated with initial pursuit acceleration, encodes importin 8, which has a key role in nuclear–cytoplasmic transport of proteins including many miRNAs.58 Importin 8 has also been identified as a component of miRNA-guided regulatory pathways for gene silencing by argonaute proteins, which are ubiquitous proteins found in plants, animals and fungi, leading to mRNA destabilization by transcription repression and translation inhibition.59 Blocking importin 8 reduces the nuclear concentration of argonaute proteins and may thus attenuate mRNA destabilization.59 We found this mutation, to date, only identified in those of AA, specifically associated with pursuit acceleration in AA probands.
Other suggestive associations with initial sensorimotor processing in the whole sample included a non-coding RNA gene (chr2p12), and SNPs ~15 kb upstream of MAP3K1 (chr5q11.2), which encodes a mitogen-activated protein kinase known to regulate apoptosis.60 There were 52 other SNPs in or around MAP3K1 including others upstream of the transcription starts site and two missense variants within the coding region, all with association P-values ranging from 3.4 × 10−5 to 2.2 × 10−7. In addition, expression quantitative trait loci analysis of the top associated SNP (rs1862618) revealed a strong correlation with the expression of the SET domain containing 9 gene (SETD9) (www.gtexportal.org/home) at chromosome 5q11.2, coding for a SET7 class of methyltransferase, which methylates H3K4. This correlation exists across multiple tissue types including regions of the brain and skeletal muscle.
Stratified GWAS by ancestry revealed significant genome-wide associations of sensorimotor processing with a synonymous mutation in Protocadherin 12 (PCDH12) in AA participants, primarily driven by effects observed in AA probands. PCDH12 belongs to a protocadherin gene cluster at chromosome 5q31 that has been previously implicated in schizophrenia and psychosis in non-AA samples.61, 62 PCDH12 encodes a cellular adhesion molecule that has an important role in cell–cell interactions including axonal guidance and synaptic specificity. The association with initial pursuit acceleration suggests that in psychotic disorders alterations of the cadherin-based adhesive system may alter functional connectivity and coherent information processing in brain systems needed for fast visual information processing.63 Putative association of PCDH12 with gyrification asymmetry has also been reported in schizophrenia suggesting its involvement in neurodevelopment and neural network formation.64 More broadly, our sensorimotor processsing related findings are in line with reports from the B-SNIP sample showing associations between genetic variants of the cadherin family and electroencephalogy abnormalities65, 66 and resting state brain activity seen with imaging studies.67
Suggestive associations of rapid sensorimotor processing around the Neurensin 1 gene (NRSN1, chr6p22.3) were observed in AA participants. NRSN1 has been suggested to have an important role in the transduction of nerve signals and for neural plasticity. This may explain why NRSN1 has been previously related to information-processing speed,68 supporting our finding of a specific association with rapid sensorimotor transformation needed during pursuit initiation.
Another genome-wide association was found for a missense mutation in the LMO7 gene coding for LIM domain only protein 7 (chr13q22.2) in AA participants in general, and in AA probands specifically at a genome-wide significant level. LMO7 is involved in protein–protein interactions and transcription, and mutations by alternative splicing in LMO7 have been related to retinal defects and degeneration,69 which could explain why we found a SNP in this gene to be associated with rapid retinal error information processing.
Stratified GWAS in CA participants revealed genome-wide significant association of sensorimotor processing with CYB5R3 (ch22q13.2). Notably, patients with 22q13 deletion syndrome are characterized by autism and schizophrenia-like symptoms.70 In these patients, loss of CYB5R3 has been related to impaired language skills.70
Genetic alterations related to pursuit maintenance
In contrast to pursuit initiation, sustained pursuit maintenance depends upon an established prediction of target velocity, and thus is more dependent on cognitive function. Here across all participants, we found suggestive associations of sustained pursuit maintenance with a region in the SH3GL2 gene (chr9p22.2) encoding Endophilin A1.71 Previous studies in schizophrenia suggest that SH3GL2 is differentially expressed in gray matter of prefrontal cortex in psychosis patients compared to controls.72, 73 Endophilin A1 is implicated in synaptic vesicle endocytosis involving intracellular signaling, calcium homeostasis and neurotransmitter release.73 Specifically, Endophilin A1 is suggested to regulate glutamate release in neurons expressing vesicular glutamate transporter 1.74 This is of interest as we recently found pursuit maintenance being associated with genetic variants in GRM3.40, 75
Genetic alterations related to antisaccade response inhibition
SNPs associated with antisaccade performance in AA participants were identified in an intergenic region at chromosome 7 with the closest defined gene being the non-coding RNA LOC101928283, which is ~230 kb away. An expression quantitative trait loci analysis search for the top 10 SNPs within the United Kingdom Brain Expression Consortium (UKBEC) (www.braineac.org) showed significant association with expression of the gene GPR37 (G protein-coupled receptor 37) within the hippocampus. The encoded protein contains seven transmembrane domains and is found in cell and endoplasmic reticulum membranes. G protein-coupled receptors are involved in translating outside signals into G protein-mediated intracellular effects. A previous GWAS on antisaccade error rate in twins reported suggestive associations with SNPs at chromosome 7 close to the region identified in the present study.76 The same study also revealed genome-wide significant associations with genes at chromosome 2.76 Others reported genome-wide linkage of antisaccade error rate with SNPs at chromosome 3p12 from a schizophrenia family study (COGS).57 Altogether, these findings support the notion that antisaccade error rate may be regarded as a complex polygenic trait.76
Implications from pathway analyses
The 200 top SNPs associated with pursuit acceleration and maintenance gain were enriched for genes related to nervous system development pathways including relevant functions such as eye formation, neuronal action potential and movement disorders. Some of these genes have also been identified in previous disease risk association studies for psychotic disorders. Altogether, these findings support the model of smooth pursuit disturbances representing alterations in brain systems contributing to psychosis disease pathology. They are in line with other findings from the B-SNIP sample that revealed brain system changes related to gene clusters indicating physiological pathways involved in brain development, synaptic transmission and ion channel activity.67, 77
Limitations
Although our findings are novel and potentially heuristically valuable, there are potential limitations. First, although our sample size was large compared to most previous association studies of eye movements in psychosis probands, it is still small for GWAS. To enhance statistical power, we used a combined proband-control sample from the B-SNIP study, which had the benefit of increasing sample size as well as phenotypic variance for genetic association analyses. However, our analyses are not powered to detect smaller genotype–phenotype associations in the individual proband groups. Further research is needed to examine potential disorder-specific effects. Second, some of our more highly associated SNP findings represented those with lower minor allele frequencies (that is, IPO8, PCDH12, CYB3R5, LMO7). Special effort was undertaken to assure SNP genotyping quality and phenotyping for these variants, however these associations require validation and replication, especially with respect to the findings in the subgroup of AA participants.
Conclusions
GWAS using eye movement phenotypes offers a promising approach for advancing pathophysiological models and understanding discrete components of complex multifactorial genetic risks for psychosis. We identified regions of interest for further study including some novel findings in addition to suggestive associations that are consistent with prior disease risk studies. Collectively, these findings highlight the importance of genes related to disease risk alongside other unique genetic contributions to eye movement phenotypes associated with psychotic disorders.
References
Thaker GK . Neurophysiological endophenotypes across bipolar and schizophrenia psychosis. Schizophr Bull 2008; 34: 760–773.
Levy DL, Sereno AB, Gooding DC, O'Driscoll GA . Eye tracking dysfunction in schizophrenia: characterization and pathophysiology. Curr Top Behav Neurosci 2010; 4: 311–347.
McDowell JE, Myles-Worsley M, Coon H, Byerley W, Clementz BA . Measuring liability for schizophrenia using optimized antisaccade stimulus parameters. Psychophysiology 1999; 36: 138–141.
Hutton SB, Ettinger U . The antisaccade task as a research tool in psychopathology: a critical review. Psychophysiology 2006; 43: 302–313.
Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD et al. Identification of distinct psychosis biotypes using brain-based biomarkers. Am J Psychiatry 2016; 173: 373–384.
Ivleva E, Thaker G, Tamminga CA . Comparing genes and phenomenology in the major psychoses: schizophrenia and bipolar 1 disorder. Schizophr Bull 2008; 34: 734–742.
Cardno AG, Owen MJ . Genetic relationships between schizophrenia, bipolar disorder, and schizoaffective disorder. Schizophr Bull 2014; 40: 504–515.
Forstner AJ, Hecker J, Hofmann A, Maaser A, Reinbold CS, Muhleisen TW et al. Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS ONE 2017; 12: e0171595.
Schulze TG, Akula N, Breuer R, Steele J, Nalls MA, Singleton AB et al. Molecular genetic overlap in bipolar disorder, schizophrenia, and major depressive disorder. World J Biol Psychiatry 2014; 15: 200–208.
Lencer R, Malchow CP, Trillenberg-Krecker K, Schwinger E, Arolt V . Eye-tracking dysfunction (ETD) in families with sporadic and familial schizophrenia. Biol Psychiatry 2000; 47: 391–401.
Ivleva EI, Moates AF, Hamm JP, Bernstein IH, O'Neill HB, Cole D et al. Smooth pursuit eye movement, prepulse inhibition, and auditory paired stimuli processing endophenotypes across the schizophrenia-bipolar disorder psychosis dimension. Schizophr Bull 2014; 40: 642–652.
Calkins ME, Iacono WG, Ones DS . Eye movement dysfunction in first-degree relatives of patients with schizophrenia: a meta-analytic evaluation of candidate endophenotypes. Brain Cogn 2008; 68: 436–461.
Hong LE, Mitchell BD, Avila MT, Adami H, McMahon RP, Thaker GK . Familial aggregation of eye-tracking endophenotypes in families of schizophrenic patients. Arch Gen Psychiatry 2006; 63: 259–264.
Clementz BA, Sweeney JA, Hirt M, Haas G . Phenotypic correlations between oculomotor functioning and schizophrenia-related characteristics in relatives of schizophrenic probands. Psychophysiology 1991; 28: 570–578.
Lencer R, Keedy SK, Reilly JL, McDonough BE, Harris MS, Sprenger A et al. Altered transfer of visual motion information to parietal association cortex in untreated first-episode psychosis: Implications for pursuit eye tracking. Psychiatry Res 2011; 194: 30–38.
Lencer R, Nagel M, Sprenger A, Heide W, Binkofski F . Reduced neuronal activity in the V5 complex underlies smooth-pursuit deficit in schizophrenia: evidence from an fMRI study. Neuroimage 2005; 24: 1256–1259.
Lencer R, Reilly JL, Harris MS, Sprenger A, Keshavan MS, Sweeney JA . Sensorimotor transformation deficits for smooth pursuit in first-episode affective psychoses and schizophrenia. Biol Psychiatry 2010; 67: 217–223.
Sweeney JA, Clementz BA, Haas GL, Escobar MD, Drake K, Frances AJ . Eye tracking dysfunction in schizophrenia: characterization of component eye movement abnormalities, diagnostic specificity, and the role of attention. J Abnorm Psychol 1994; 103: 222–230.
Sweeney JA, Luna B, Srinivasagam NM, Keshavan MS, Schooler NR, Haas GL et al. Eye tracking abnormalities in schizophrenia: evidence for dysfunction in the frontal eye fields. Biol Psychiatry 1998; 44: 698–708.
Sweeney JA, Luna B, Haas GL, Keshavan MS, Mann JJ, Thase ME . Pursuit tracking impairments in schizophrenia and mood disorders: step-ramp studies with unmedicated patients. Biol Psychiatry 1999; 46: 671–680.
Trillenberg P, Sprenger A, Talamo S, Herold K, Helmchen C, Verleger R et al. Visual and non-visual motion information processing during pursuit eye tracking in schizophrenia and bipolar disorder. Eur Arch Psychiatry Clin Neurosci 2017; 267: 225–235.
O'Driscoll GA, Alpert NM, Matthysse SW, Levy DL, Rauch SL, Holzman PS . Functional neuroanatomy of antisaccade eye movements investigated with positron emission tomography. Proc Natl Acad Sci USA 1995; 92: 925–929.
O'Driscoll GA, Benkelfat C, Florencio PS, Wolff AL, Joober R, Lal S et al. Neural correlates of eye tracking deficits in first-degree relatives of schizophrenic patients: a positron emission tomography study. Arch Gen Psychiatry 1999; 56: 1127–1134.
McDowell JE, Brown GG, Paulus M, Martinez A, Stewart SE, Dubowitz DJ et al. Neural correlates of refixation saccades and antisaccades in normal and schizophrenia subjects. Biol Psychiatry 2002; 51: 216–223.
Ettinger U, Kumari V, Chitnis XA, Corr PJ, Crawford TJ, Fannon DG et al. Volumetric neural correlates of antisaccade eye movements in first-episode psychosis. Am J Psychiatry 2004; 161: 1918–1921.
Tu P, Buckner RL, Zollei L, Dyckman KA, Goff DC, Manoach DS . Reduced functional connectivity in a right-hemisphere network for volitional ocular motor control in schizophrenia. Brain 2010; 133: 625–637.
Hong LE, Turano KA, O'Neill H, Hao L, Wonodi I, McMahon RP et al. Refining the predictive pursuit endophenotype in schizophrenia. Biol Psychiatry 2008; 63: 458–464.
Reilly JL, Frankovich K, Hill S, Gershon ES, Keefe RS, Keshavan MS et al. Elevated antisaccade error rate as an intermediate phenotype for psychosis across diagnostic categories. Schizophr Bull 2014; 40: 1011–1021.
Lencer R, Sprenger A, Reilly JL, McDowell JE, Rubin LH, Badner JA et al. Pursuit eye movements as an intermediate phenotype across psychotic disorders: evidence from the B-SNIP study. Schizophr Res 2015; 169: 326–333.
Tamminga CA, Ivleva EI, Keshavan MS, Pearlson GD, Clementz BA, Witte B et al. Clinical phenotypes of psychosis in the bipolar-schizophrenia network on intermediate phenotypes (B-SNIP). Am J Psychiatry 2013; 170: 1263–1274.
Arolt V, Lencer R, Nolte A, Muller-Myhsok B, Purmann S, Schurmann M et al. Eye tracking dysfunction is a putative phenotypic susceptibility marker of schizophrenia and maps to a locus on chromosome 6p in families with multiple occurrence of the disease. Am J Med Genet 1996; 67: 564–579.
Matthysse S, Holzman PS, Gusella JF, Levy DL, Harte CB, Jorgensen A et al. Linkage of eye movement dysfunction to chromosome 6p in schizophrenia: additional evidence. Am J Med Genet 2004; 128B: 30–36.
Haraldsson HM, Ettinger U, Magnusdottir BB, Sigmundsson T, Sigurdsson E, Ingason A et al. COMT val(158)met genotype and smooth pursuit eye movements in schizophrenia. Psychiatry Res 2009; 169: 173–175.
Park BL, Shin HD, Cheong HS, Park CS, Sohn JW, Kim BJ et al. Association analysis of COMT polymorphisms with schizophrenia and smooth pursuit eye movement abnormality. J Hum Genet 2009; 54: 709–712.
Rybakowski JK, Borkowska A, Czerski PM, Hauser J . Eye movement disturbances in schizophrenia and a polymorphism of catechol-O-methyltransferase gene. Psychiatry Res 2002; 113: 49–57.
Haraldsson HM, Ettinger U, Magnusdottir BB, Ingason A, Hutton SB, Sigmundsson T et al. Neuregulin-1 genotypes and eye movements in schizophrenia. Eur Arch Psychiatry Clin Neurosci 2010; 260: 77–85.
Smyrnis N, Kattoulas E, Stefanis NC, Avramopoulos D, Stefanis CN, Evdokimidis I . Schizophrenia-related neuregulin-1 single-nucleotide polymorphisms lead to deficient smooth eye pursuit in a large sample of young men. Schizophr Bull 2011; 37: 822–831.
Schmechtig A, Vassos E, Kumari V, Hutton SB, Collier DA, Morris RG et al. Association of neuregulin 1 rs3924999 genotype with antisaccades and smooth pursuit eye movements. Genes Brain Behav 2010; 9: 621–627.
Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ et al. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry 2011; 168: 930–946.
Lencer R, Bishop JR, Harris MS, Reilly JL, Patel S, Kittles R et al. Association of variants in DRD2 and GRM3 with motor and cognitive function in first-episode psychosis. Eur Arch Psychiatry Clin Neurosci 2014; 264: 345–355.
First MB, Spitzer RL, Gibbon M, Williams JBW . Structured Clinical Interview for DSM-IV Axis I Disorders, Patient Edition (SCID-P). New York State Psychiatric Institute: New York, NY, USA, 1995.
Wilkinson GS, Robertson GJ . Psychological Assessment Resources I. WRAT 4: Wide Range Achievement Test, 4th edn. Psychological Assessment Resources: Lutz, FL, USA, 2006.
Keefe RS, Harvey PD, Goldberg TE, Gold JM, Walker TM, Kennel C et al. Norms and standardization of the brief assessment of cognition in schizophrenia (BACS). Schizophr Res 2008; 102: 108–115.
Kay SR, Fiszbein A, Opler LA . The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull 1987; 13: 261–276.
Montgomery SA, Asberg M . A new depression scale designed to be sensitive to change. Br J Psychiatry 1979; 134: 382–389.
Young RC, Biggs JT, Ziegler VE, Meyer DA . A rating scale for mania: reliability, validity and sensitivity. Br J Psychiatry 1978; 133: 429–435.
Andreasen NC, Pressler M, Nopoulos P, Miller D, Ho BC . Antipsychotic dose equivalents and dose-years: a standardized method for comparing exposure to different drugs. Biol Psychiatry 2010; 67: 255–262.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT . Data quality control in genetic case-control association studies. Nat Protoc 2010; 5: 1564–1573.
Sun L, Dimitromanolakis A . PREST-plus identifies pedigree errors and cryptic relatedness in the GAW18 sample using genome-wide SNP data. BMC Proc 2014; 8: S23.
Williams AL, Patterson N, Glessner J, Hakonarson H, Reich D . Phasing of many thousands of genotyped samples. Am J Hum Genet 2012; 91: 238–251.
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR . Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–959.
Genomes Project Consortium Genomes Project Consortium Auton A, Genomes Project Consortium Brooks LD, Genomes Project Consortium Durbin RM, Genomes Project Consortium Garrison EP, Genomes Project Consortium Kang HM et al. A global reference for human genetic variation. Nature 2015; 526: 68–74.
Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet 2010; 42: 348–354.
Li MX, Yeung JM, Cherny SS, Sham PC . Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet 2012; 131: 747–756.
Quinlan AR, BEDTools . The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 2014; 47: 11 12 11–11 12 34.
Greenwood TA, Swerdlow NR, Gur RE, Cadenhead KS, Calkins ME, Dobie DJ et al. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry 2013; 170: 521–532.
Wei Y, Li L, Wang D, Zhang CY, Zen K . Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem 2014; 289: 10270–10275.
Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P et al. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 2009; 136: 496–507.
Schlesinger TK, Bonvin C, Jarpe MB, Fanger GR, Cardinaux JR, Johnson GL et al. Apoptosis stimulated by the 91-kDa caspase cleavage MEKK1 fragment requires translocation to soluble cellular compartments. J Biol Chem 2002; 277: 10283–10291.
Schwab SG, Eckstein GN, Hallmayer J, Lerer B, Albus M, Borrmann M et al. Evidence suggestive of a locus on chromosome 5q31 contributing to susceptibility for schizophrenia in German and Israeli families by multipoint affected sib-pair linkage analysis. Mol Psychiatry 1997; 2: 156–160.
Lachman HM, Petruolo OA, Pedrosa E, Novak T, Nolan K, Stopkova P . Analysis of protocadherin alpha gene deletion variant in bipolar disorder and schizophrenia. Psychiatr Genet 2008; 18: 110–115.
Redies C, Hertel N, Hubner CA . Cadherins and neuropsychiatric disorders. Brain Res 2012; 1470: 130–144.
Gregorio SP, Sallet PC, Do KA, Lin E, Gattaz WF, Dias-Neto E . Polymorphisms in genes involved in neurodevelopment may be associated with altered brain morphology in schizophrenia: preliminary evidence. Psychiatry Res 2009; 165: 1–9.
Mokhtari M, Narayanan B, Hamm JP, Soh P, Calhoun VD, Ruano G et al. Multivariate genetic correlates of the auditory paired stimuli-based P2 event-related potential in the psychosis dimension from the BSNIP study. Schizophr Bull 2016; 42: 851–862.
Narayanan B, Ethridge LE, O'Neil K, Dunn S, Mathew I, Tandon N et al. Genetic sources of subcomponents of event-related potential in the dimension of psychosis analyzed from the B-SNIP study. Am J Psychiatry 2015; 172: 466–478.
Meda SA, Ruano G, Windemuth A, O'Neil K, Berwise C, Dunn SM et al. Multivariate analysis reveals genetic associations of the resting default mode network in psychotic bipolar disorder and schizophrenia. Proc Natl Acad Sci USA 2014; 111: E2066–E2075.
Luciano M, Hansell NK, Lahti J, Davies G, Medland SE, Raikkonen K et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol 2011; 86: 193–202.
Putilina T, Jaworski C, Gentleman S, McDonald B, Kadiri M, Wong P . Analysis of a human cDNA containing a tissue-specific alternatively spliced LIM domain. Biochem Biophys Res Commun 1998; 252: 433–439.
Sarasua SM, Dwivedi A, Boccuto L, Chen CF, Sharp JL, Rollins JD et al. 22q13.2q13.32 genomic regions associated with severity of speech delay, developmental delay, and physical features in Phelan-McDermid syndrome. Genet Med 2014; 16: 318–328.
Giachino C, Lantelme E, Lanzetti L, Saccone S, Bella Valle G, Migone N . A novel SH3-containing human gene family preferentially expressed in the central nervous system. Genomics 1997; 41: 427–434.
Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry 2004; 9: 684–697.
Martins-de-Souza D, Gattaz WF, Schmitt A, Rewerts C, Maccarrone G, Dias-Neto E et al. Prefrontal cortex shotgun proteome analysis reveals altered calcium homeostasis and immune system imbalance in schizophrenia. Eur Arch Psychiatry Clin Neurosci 2009; 259: 151–163.
Weston MC, Nehring RB, Wojcik SM, Rosenmund C . Interplay between VGLUT isoforms and endophilin A1 regulates neurotransmitter release and short-term plasticity. Neuron 2011; 69: 1147–1159.
Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci USA 2004; 101: 12604–12609.
Vaidyanathan U, Malone SM, Donnelly JM, Hammer MA, Miller MB, McGue M et al. Heritability and molecular genetic basis of antisaccade eye tracking error rate: a genome-wide association study. Psychophysiology 2014; 51: 1272–1284.
Tandon N, Nanda P, Padmanabhan JL, Mathew IT, Eack SM, Narayanan B et al. Novel gene-brain structure relationships in psychotic disorder revealed using parallel independent component analyses. Schizophr Res 2017; 182: 74–83.
Acknowledgements
We thank the study participants who contributed their time and effort to participate in this study. We also thank Gunvant Thaker, MD, for his many scientific contributions to the B-SNIP consortium especially in regards to the eye movement studies. This work was supported by the National Institute of Mental Health (Grant Numbers MH077851 (CAT), MH078113 (MSK), MH077945 (GDP), MH077852 (GKT), MH085485 (BAC), MH077862 (JAS) and MH083888 (JRB)). This work was further supported by the Alexander von Humboldt Foundation, Germany (to RL and JS).
Disclaimer
The NIMH had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or the decision to submit the manuscript for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
CAT reports the following financial disclosures: American Psychiatric Association, Deputy Editor; Astellas, Ad Hoc Consultant; Autifony, Ad Hoc Consultant; The Brain and Behavior Foundation, Council Member; Eli Lilly Pharmaceuticals, Ad Hoc Consultant; Intra-cellular Therapies (ITI), Advisory Board, drug development; Institute of Medicine, Council Member; National Academy of Medicine, Council Member; Pfizer, Ad Hoc Consultant; Sunovion, Investigator Initiated grant funding. JAS reports the following financial disclosures: ad hoc consultant to Takeda Pharmaceuticals. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lencer, R., Mills, L., Alliey-Rodriguez, N. et al. Genome-wide association studies of smooth pursuit and antisaccade eye movements in psychotic disorders: findings from the B-SNIP study. Transl Psychiatry 7, e1249 (2017). https://doi.org/10.1038/tp.2017.210
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2017.210
This article is cited by
-
Investigation of genetic determinants of cognitive change in later life
Translational Psychiatry (2024)
-
Genome-wide association studies and cross-population meta-analyses investigating short and long sleep duration
Nature Communications (2023)
-
Expression Analysis of Ermin and Listerin E3 Ubiquitin Protein Ligase 1 Genes in the Periphery of Patients with Schizophrenia
Journal of Molecular Neuroscience (2022)
-
Multivariate relationships between peripheral inflammatory marker subtypes and cognitive and brain structural measures in psychosis
Molecular Psychiatry (2021)
-
Eye movements in patients in early psychosis with and without a history of cannabis use
npj Schizophrenia (2021)