Abstract
Intranasal administration of the neuropeptide oxytocin has been shown to influence a range of complex social cognitions and social behaviors, and it holds therapeutic potential for the treatment of mental disorders characterized by social functioning deficits such as autism, social phobia and borderline personality disorder. However, considerable variability exists in individual responses to oxytocin administration. Here, we undertook a study to investigate the role of genetic variation in sensitivity to exogenous oxytocin using a socioemotional task. In a randomized, double-blind, placebo-controlled experiment with a repeated-measures (crossover) design, we assessed the performance of 203 men on an emotion recognition task under oxytocin and placebo. We took a haplotype-based approach to investigate the association between oxytocin receptor gene variation and oxytocin sensitivity. We identified a six-marker haplotype block spanning the promoter region and intron 3 that was significantly associated with our measure of oxytocin sensitivity. Specifically, the TTCGGG haplotype comprising single-nucleotide polymorphisms rs237917–rs2268498–rs4564970–rs237897–rs2268495–rs53576 is associated with increased emotion recognition performance under oxytocin versus placebo, and the CCGAGA haplotype with the opposite pattern. These results on the genetic modulation of sensitivity to oxytocin document a significant source of individual differences with implications for personalized treatment approaches using oxytocin administration.
Similar content being viewed by others
Introduction
The neuropeptide oxytocin has a key role in the regulation of complex social cognition and behaviors, including mammalian pair bonding, maternal behavior, recognition of conspecifics and responses to social stress.1, 2 Human studies have documented a number of effects of intranasally administered oxytocin on social behavior and social cognition.3, 4, 5 For instance, a single dose of intranasal oxytocin increases trusting behavior,6 attention to the eye region of faces7, 8 and the accuracy with which individuals interpret emotional information from eyes.9 Oxytocin also reduces subjective and physiological responses to social stress and enhances the stress-buffering effect of social support.10 Neuroimaging studies have shown that one of the mechanisms through which oxytocin influences social cognition is by attenuating amygdala responses to social stimuli such as emotional faces.11, 12, 13, 14
There has also been considerable interest in the therapeutic potential of oxytocin for the treatment of mental disorders characterized by social functioning deficits such as autism,15, 16, 17, 18, 19 social phobia20, 21 and borderline personality disorder.22 Whereas the results of virtually all of these studies suggest that oxytocin has the potential to improve social cognition and social behavior in healthy individuals and patient populations, there is considerable variability in individual responsiveness to oxytocin administration, with some individuals showing strong behavioral effects in response to intranasal oxytocin and others seeming to show minimal or no effects.3 These findings suggest that oxytocin administration may interact with pre-existing interindividual variation that influences the efficacy of oxytocin signaling in the central nervous system. Specifically, genetic variation influencing the number, distribution or functioning of oxytocin receptors may influence oxytocin sensitivity. Research investigating the genetic modulation of sensitivity to oxytocin administration has implications not only for basic research but also implications for clinicians who would like to predict which patients may or may not benefit from oxytocin administration, and in general may help to refine personalized oxytocin-based therapies for mental disorders.
Initial studies using a molecular genetic approach suggest that common variations in the human oxytocin system are relevant for social behavior.23 Specifically, variation in the human oxytocin receptor gene (OXTR) has been associated in some studies to susceptibility for mental disorders characterized by social deficits such as autism spectrum disorder24 (although the specific single-nucleotide polymorphism(s) (SNP(s)) and allele(s) linked to greater susceptibility have varied depending on ethnicity and other sample characteristics).25, 26, 27, 28 In addition, variants of OXTR have been linked to individual differences in psychological and physiological response patterns to stress and social information,29, 30 as well as to differences in general sociobehavioral phenotypes. These include prosocial behavior,31 empathy,32 trust33 and sensitive parenting.34 Structural and functional imaging studies have shown associations between OXTR SNPs and morphometric alterations as well as differences in activity of neural circuits involved in the processing of social information and negative affect. These imaging studies suggest that genetic variation of OXTR influences limbic circuitry involving the amygdala, the hypothalamus and the cingulate gyrus.35, 36, 37, 38
Given the associations that have been documented between OXTR variability and social behavior, and given that oxytocin exerts its function via the oxytocin receptor, OXTR variability is a prime candidate for explaining individual differences in response to exogenous oxytocin administration. To test this possibility, we conducted a randomized, double-blind, placebo-controlled experiment with a repeated-measures (crossover) design. We assessed each participant’s performance on an emotion recognition task previously shown to be sensitive to oxytocin administration,39 both under oxytocin and placebo. This within-subject design provides a measure of each individual’s socioemotional sensitivity to oxytocin administration.
Whereas most association studies so far have focused on single SNPs, we investigated 23 SNPs across the OXTR gene region and took a haplotype-based approach in our analyses. In addition to allowing us to capture more genetic information than single SNP analysis,40 the haplotype approach—which by definition jointly tests multiple SNPs for association—allows us to avoid the conundrum of multiple testing, which can result in false-positive results. As has been noted by other researchers,41 divergent results between studies may sometimes have arisen from unrecognized haplotype effects because these studies were limited to investigation of individual SNPs. In addition, questionable results may arise from disregarding linkage disequilibrium (LD) of the tested polymorphism, an effect preventable by haplotype analysis (see Binder et al.42 for further discussion of haplotype analysis).
Materials and methods
Subjects
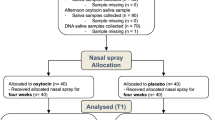
We tested 207 men between 19 and 30 years of age (M=23.5 years, s.d.=2.7 years) of Central European descent from the student population of the University of Freiburg, Germany, who were recruited through on-campus advertisements. The sample size was determined on the basis of recent research demonstrating effects of common variants of OXTR on behavior.43, 44, 45, 46, 47 Participants were pre-screened to ensure that they were fluent in German, were not suffering from mental or physical illnesses and were not taking drugs or medication. Students studying psychology were excluded from our sample. All participants gave written, informed consent before the start of the first experimental session and received a total of 40 Euro for participation in the two sessions. The study was approved by the institutional review board of the University of Freiburg.
Procedure
Each participant was tested twice, with a 1-week gap between testing sessions. All sessions took place in the afternoon and lasted ~2 h. Participants were tested in a laboratory in groups of 12–16, with each participant seated at a computer in an individual cubicle. Dividing walls prevented visual contact between the participants, and participants were instructed not to speak to each other during the testing sessions.
Substance administration
After completing a questionnaire to assess baseline mood (the Multidimensional Mood Questionnaire48), participants received either oxytocin (Syntocinon-Spray, Novartis, Basel, Switzerland) or placebo intranasally in a randomized, double-blind, crossover design. The placebo included all of the same ingredients as the oxytocin spray except for the neuropeptide. Under the supervision of the experimenter, participants self-administered 24 IU of the substance (three puffs into each nostril in alternating order).10 After a loading period of 45 min, the participants completed a second Multidimensional Mood Questionnaire to assess their mood.
Emotion recognition task
Participants subsequently completed a task based closely on one used by Lischke et al.39 to assess their ability to recognize emotions from dynamic facial expressions. Previous research suggests that this task is sensitive to oxytocin administration: participants who received oxytocin identified emotions more quickly and at a lower intensity level than participants who received placebo.39 In each trial, a photo of an individual displaying a neutral expression gradually morphed into a photo of the same individual displaying a happy, angry, fearful or sad expression. The participant’s task was to press a button when he recognized the emotion being displayed. Participants were instructed to respond as quickly and accurately as possible. As soon as the participant responded, the morphing procedure stopped and the image was replaced by a prompt asking the participant to indicate which of the four emotions he had detected (see Supplementary Information for details).
Non-social control task
To test whether any observed effects would be specific to social stimuli, participants also completed a control task for the recognition of non-social stimuli (cars) after the emotion recognition task. Using Winmorph 3.01 (http://www.debugmode.com/winmorph/), grayscale images of a common car model (a small hatchback) were morphed into images of target cars (four exemplars each of the car types: Smart car, box truck, pickup truck and VW Beetle). As in the emotion recognition task, participants were instructed to press a button as soon as they recognized the target car type. Before the 16 test trials, participants completed four practice trials. The test trials appeared in one of two pseudorandomized testing orders. For each trial, reaction time and accuracy were recorded.
During the second session, a saliva sample was collected from each participant (Oragene 500-OG kit, DNA Genotek, Ottawa, Ontario, Canada) for genotyping. Participants were also asked at the end of the second session whether they thought they had received oxytocin or placebo in that session.
SNP selection and genotyping
The goal of this study was to associate variation of OXTR with oxytocin sensitivity. In order to capture the maximum amount of genetic information, we chose 23 SNPs across the OXTR gene for subsequent haplotype-based analyses. Sixteen SNPs were selected on the basis of an association study with SNP coverage across OXTR.26 Seven additional SNPs were selected; rs53576 was included because a large number of studies have shown significant associations between this SNP and sociobehavioral phenotypes.23 SNP rs2268498, located in the promoter region, was included because of its presumed functional significance.49 In order to increase coverage of the 5′ region, a likely region of regulatory activity, four additional SNPs compatible with the multiplex assay were selected from SNP databases. This selection procedure is a standard a priori approach to haplotype testing, where not a single SNP but rather SNPs across a gene region and their pattern of LD are taken into account.
DNA was extracted from saliva in Oragene collection vials by desalting procedure following the manufacturer’s protocol. SNP genotyping was performed on the Sequenom MassArray platform using iPLEX Gold chemistry (Sequenom, San Diego, CA, USA). Primers and multiplexes were designed using the Sequenom's MassArray Assay Design v4.0 Software. Multiplex PCR was performed using iPlex Gold reagents in 5-μl reactions as recommended by the manufacturer. Three hundred eighty-four-well PCR plates were used for PCR amplification on a GeneAmp 9700 thermocycler (Applied Biosystems, Waltham, MA, USA), and the products were spotted onto 384-well SpectroCHIP arrays (Sequenom) using the Nanodispenser RS1000 (Sequenom). Spotted SpectroCHIPs were then fired in a MassArray Analyzer 4 Matrix-Assisted Laser Desorption/Ionization-Time Of Flight mass spectrometer. Procedures used were as in the manufacturer's protocol. Data analyses were performed on Sequenom's Typer Software Module, and ambiguous genotype calls were manually confirmed. Out of the 23 genotyped SNPs (see Supplementary Table 1), 7 were not in Hardy–Weinberg equilibrium and 2 SNPs had a minor allele frequency below 5%. Genotyping failed for SNP rs237894. Therefore, 13 SNPs were included in the association test (see Results).
Statistics
We analyze data from all participants (N=203) who completed the emotion recognition task in both sessions and for whom genotyping was successful. Accuracy of emotion recognition and reaction time were recorded for each trial. Accuracy was near ceiling (M=88%, s.d.=9%) and is thus not considered in the following analyses. Reaction times were averaged across all accurate trials.
Preliminary analyses showed a main effect of time, indicating that reaction times improved between the first and second sessions for all groups. Therefore, reaction times were standardized (z-transformed) separately for each session (1 and 2). In order to obtain a measure for oxytocin sensitivity, a difference score was calculated (subtracting standardized reaction times under oxytocin from standardized reaction times under placebo).
Haplotype analyses were performed with version 3.1.6 of the program UNPHASED.50 Associations between haplotypes and the quantitative trait oxytocin sensitivity were tested with the individual haplotype test implemented in UNPHASED. This option gives individual tests for each haplotype in turn, and tests for a difference between a haplotype and all the others pooled together. The outcome variable was our measure for oxytocin sensitivity, that is, the difference in reaction time between oxytocin and placebo. Using a sliding window approach, seven to two marker haplotypes were tested. Only haplotypes with a frequency of >2.5% were analyzed. In order to correct for multiple testing, the permutation test option as provided in UNPHASED was used. Permutation test correction was performed using 1000 iterations (random permutations) and was applied to correction of global P-values (the P-value generated by UNPHASED as an overall test of each block of haplotypes).
In order to confirm the results obtained using UNPHASED, which used the difference score as an outcome measure, a General Linear Model for repeated measures, with reaction time under oxytocin and placebo as repeated variables, haplotype as the between-subject factor, and order of substance administration as a covariate was computed (SPSS Version 20, IBM, Armonk, NY, USA). Assumptions of normality, equality of variance in the groups being compared, and sphericity were checked and met by the data. In all analyses, two-tailed P-values are reported.
Results
The assayed SNPs, their location along the OXTR genomic region and their relationship to the exon/intron boundaries of the gene are shown in the upper part of Figure 1. The lower part shows the LD map of the genotyped SNPs as depicted by Haploview.
Haplotype analyses revealed significant associations between oxytocin sensitivity and haplotypes of marker sizes six to two (Table 1). A more detailed analysis of the six-marker block showed that two haplotypes differed significantly from the remaining ones: CCGAGA and TTCGGG (with frequencies of 7.1% and 28.3%, respectively; see Table 2).
In order to specify the direction of effects, we compared oxytocin sensitivity scores (based on the emotion recognition speed under oxytocin versus placebo; see Materials and methods) among three groups: carriers of the two significant haplotype groups (TTCGGG and CCGAGA) and the group comprising the remaining haplotype combinations (Other group). As shown in Figure 2, the two significant six-marker haplotypes showed opposing effects. Both carriers of the TTCGGG haplotype and carriers of the CCGAGA haplotype showed differential sensitivity to oxytocin relative to the Other group. Specifically, carriers of the TTCGGG haplotype recognized emotions more quickly and at a lower intensity level under oxytocin compared with placebo; CCGAGA carriers showed the opposite pattern. When all genotype groups were combined, there was no main effect of oxytocin on emotion recognition speed.
Graph shows relative oxytocin sensitivity across groups for the six-marker haplotype block rs237917–rs2268498–rs4564970–rs237897–rs2268495–rs53576. Oxytocin sensitivity was indexed in terms of emotion recognition speed under oxytocin compared with placebo. Higher values indicate faster reaction times under oxytocin.
The results obtained using UNPHASED, which used the difference score as the outcome measure, were confirmed with a general linear model for repeated measures, with reaction time under oxytocin and placebo as repeated variables, six-marker haplotype as the between-subject factor and order of substance administration as a covariate (haplotype by substance interaction: F2,186=5.80; P=0.004).
With regard to the blocks of smaller window sizes, with very few exceptions, the same allelic combinations (comprising subsets of the alleles in the six-marker haplotypes) drove the significant associations (see Supplementary Table 2; detailed results for the other haplotype windows can be found in Supplementary Table 3).
There is one additional three-marker haplotype in intron 3 comprising SNPs rs2254298–rs2268494–rs9840864 showing significant results (Figure 3). As shown in Table 3, the A-allele of rs2254298, which also shows a nominally significant association on the single SNP level, falls exclusively on the significant haplotype. Furthermore, the six SNPs that make up the significant six-marker haplotype and additionally rs2254298 were significantly associated with sensitivity to oxytocin on their own (see bottom of Table 1), with rs237897 surviving Bonferroni correction for 13 tested SNPs.
Non-social control condition and additional analyses
General Linear Model analyses showed no haplotype by substance interactions (F2,180=1.52; P=0.22) under the non-social control condition, which used cars that morphed from a ‘neutral’ model into one of four other models as stimuli, supporting the view that the effects of oxytocin are specific for social stimuli.
Finally, as a group, participants were not able to tell whether they had received oxytocin or placebo (the accuracy rate of 53% did not differ from the chance level of 50%, Z=0.85, P=0.39). Participants’ mood was not affected by receiving oxytocin versus placebo (that is, changes in Multidimensional Mood Questionnaire scores from pre- to post-spray application did not differ by the substance group, all P>0.20).
Discussion
There are several lines of evidence implicating the central oxytocin system in the etiology of mental disorders characterized by social deficits. This is supported by genetic studies linking SNPs and haplotypes of OXTR to individual differences in social behavior. Furthermore, several of these SNPs have been associated with structural and functional changes in human brain regions involved in a regulatory circuit of socioemotional information processing. The discovery that neuropeptides can be non-invasively delivered to the brain in humans has raised considerable interest in the therapeutic potential of oxytocin for the treatment of disorders characterized by social functioning deficits.3, 4
Here, we investigated whether sensitivity to oxytocin administration is influenced by genetic variation of OXTR. We identified a six-marker haplotype block spanning the promoter region and intron 3 that was significantly associated with our measure of oxytocin sensitivity, emotion recognition performance under oxytocin versus placebo. Specifically, we identified two haplotypes that were differentially associated with oxytocin sensitivity: the TTCGGG haplotype comprising SNPs rs237917–rs2268498–rs4564970–rs237897–rs2268495–rs53576 is associated with increased emotion recognition performance under oxytocin versus placebo, and the CCGAGA haplotype with the opposite pattern. It is of note that the A allele of rs53576, previously associated with phenotypes including reduced empathy, lower trust, reduced sensitivity to social support in the context of stress and less sensitive parenting, falls almost exclusively on the CCGAGA haplotype. In the present study, this haplotype was associated with decreased emotion recognition speed under oxytocin compared with placebo.
The functional consequences associated with this haplotype are currently unknown. Speculatively, it is possible that previously reported findings in the literature involving rs53576 are functionally explained by LD with the SNPs located in the regulatory region of OXTR, possibly influencing transcriptional efficiency. There is tentative evidence that promoter SNP rs2268498, associated with negative emotionality49 and moral judgments,51 influences OXTR gene regulation (referred to in Montag et al.49 as unpublished data). Detailed functional in vitro characterization of OXTR promoter SNPs, including rs2268498, is warranted.
Although a number of other studies have used single SNP approaches to examine the effect of oxytocin intranasal administration on social cognition, we are not aware of the specific use of haplotype analysis in experiments that involve intranasal oxytocin administration. There are studies that have used haplotype analysis to examine association between the oxytocin receptor, behavior and psychopathology. For instance, in a study by Lerer et al.,26 several OXTR haplotypes were associated with autism. Significant findings emerged for haplotypes comprising SNPs in intron 1 and the untranslated part of exon 4. Of note, associations were shown with haplotypes of several sizes including SNPs rs237897 and/or rs2254298. The first SNP is part of our six-marker haplotype, and rs2254298 is tagging the three-marker haplotype identified in the present study. Another haplotype-based analysis identified an association between social cognition and a haplotype consisting of rs11131149 and rs2254298.52 A third study showed that a haplotype comprising SNPs rs9840864 and rs2268494—the latter being part of the significant three-marker block in our study—was associated with anger and retaliation after betrayal.47 This shows some convergence between studies; however, as our study had a more extensive coverage of the 5′ region of the gene, and the other studies did not or only partially include the SNPs of our six-marker haplotype, the results cannot be directly compared.
Our results suggest that oxytocin administration facilitates emotion recognition in some individuals more than others. The fact that no main effect of oxytocin was observed in this study when all genotype groups were combined highlights the critical influence of individual difference factors in oxytocin administration effects. The ability to recognize the emotions of others is a building block for empathy and has been associated with prosocial behavior.53 Impairments in the ability to recognize emotions are also characteristic of several mental disorders involving social deficits, including autism54 and borderline personality disorder.55 Preliminary evidence suggests that oxytocin may indeed alleviate impairments in emotion recognition in some clinical samples.8, 18, 56 By identifying particular individuals who respond to oxytocin administration more strongly than others in the context of an emotion recognition task, the current study provides a first step toward predicting which individuals are likely to profit from oxytocin-based treatments intended to alleviate impairments in socioemotional functioning.
We chose the emotion recognition task in our study based on the fact that it has been shown to be sensitive to oxytocin administration.39 Furthermore, because it involves dynamic stimuli, this task arguably has more ecological validity than tasks involving static photos of faces. However, future research will be necessary to test the generalizability of our reported findings to other emotion recognition tasks, to other tasks relevant for empathy and prosocial behavior and to other types of settings and contexts.
Because previous results have suggested sex differences in associations between OXTR genotype and phenotype57 as well as in responses to intranasal oxytocin administration,11, 58 we decided to limit our current study to male participants. Follow-up studies testing women are necessary to examine the generalizability of our findings and are expected to be particularly informative, given the potential for sex differences in this domain. As with all genetic association studies, our findings should be replicated in an independent sample and with a larger sample size to establish the robustness of the effects we observed. Given that different directions of genotype–phenotype associations have been observed in different ethnicities,25, 28 future work should also test the effects reported here in other ethnic groups. Furthermore, the distribution of these haplotypes in samples with impairments in socioemotional functioning should be documented in future research. This information on haplotype distributions may help to elucidate some of the potential sources of these socioemotional impairments and may also help identify specific samples for which oxytocin-based therapies are more or less likely to be effective.
It is also likely that epigenetic modifications of OXTR influence oxytocin sensitivity.59 Functional studies have shown that differential methylation of a CpG island in the OXTR promoter seems to be functionally important for OXTR expression,60 and differences in the degree of methylation have been observed in childhood disorders characterized by impairments in social cognition, including autism61 and conduct problems concurrent with callous-unemotional traits.62
In addition to OXTR, other factors involved in the oxytocin pathway might influence oxytocin sensitivity. These include the gene for oxytocin (OXT; coding for the precursor protein oxytocin-neurophysin-I), the gene encoding the enzyme that metabolizes oxytocin, oxytocinase (human leucyl/cystinylaminopeptidase; LNPEP) as well as CD38, a key mediator of oxytocin brain release.63, 64 In the long term, future studies involving larger sample sizes may help to elucidate the role of these other genes, along with potential gene–gene interactions in oxytocin sensitivity.
The current study, which demonstrates genetic modulation of sensitivity to oxytocin administration, identifies one specific and systematic source of individual differences in response to oxytocin administration. The identification of this new haplotype block conferring differences in oxytocin-induced socioemotional behavior will not only help advance a basic scientific understanding of the human oxytocin system but may also improve the prediction of clinical outcomes of novel therapy approaches using oxytocin. More specifically, these results may help to bridge the insights from a pharmacogenetic approach to psychobiological therapy combining oxytocin administration and psychotherapy4, 65, 66 for genetically targeted subgroups of patients with social deficits across diagnostic categories of mental disorders (for example, autism spectrum disorder, social anxiety disorder and borderline personality disorder). More such personalized treatment strategies67, 68 are necessary to help fulfill the immense promise of translational success of oxytocin-based therapies.
References
Donaldson ZR, Young LJ . Oxytocin, vasopressin, and the neurogenetics of sociality. Science 2008; 322: 900–904.
Insel TR . The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron 2010; 65: 768–779.
Meyer-Lindenberg A, Domes G, Kirsch P, Heinrichs M . Oxytocin and vasopressin in the human brain: social neuropeptides for translational medicine. Nat Rev Neurosci 2011; 12: 524–538.
Heinrichs M, von Dawans B, Domes G . Oxytocin, vasopressin, and human social behavior. Front Neuroendocrinol 2009; 30: 548–557.
Feldman R . Oxytocin and social affiliation in humans. Hormones Behav 2012; 61: 380–391.
Kosfeld M, Heinrichs M, Zak PJ, Fischbacher U, Fehr E . Oxytocin increases trust in humans. Nature 2005; 435: 673–676.
Guastella AJ, Mitchell PB, Dadds MR . Oxytocin increases gaze to the eye region of human faces. Biol Psychiatry 2008; 63: 3–5.
Domes G, Kumbier E, Heinrichs M, Herpertz SC . Oxytocin promotes facial emotion recognition and amygdala reactivity in adults with asperger syndrome. Neuropsychopharmacology 2014; 39: 698–706.
Domes G, Heinrichs M, Michel A, Berger C, Herpertz SC . Oxytocin improves ‘mind-reading‘ in humans. Biol Psychiatry 2007; 61: 731–733.
Heinrichs M, Baumgartner T, Kirschbaum C, Ehlert U . Social support and oxytocin interact to suppress cortisol and subjective responses to psychosocial stress. Biol Psychiatry 2003; 54: 1389–1398.
Domes G, Heinrichs M, Glascher J, Buchel C, Braus D, Herpertz S . Oxytocin attenuates amygdala responses to emotional faces regardless of valence. Biol Psychiatry 2007; 62: 1187–1190.
Gamer M, Zurowski B, Buchel C . Different amygdala subregions mediate valence-related and attentional effects of oxytocin in humans. Proc Natl Acad Sci USA 2010; 107: 9400–9405.
Kirsch P, Esslinger C, Chen Q, Mier D, Lis S, Siddhanti S et al. Oxytocin modulates neural circuitry for social cognition and fear in humans. J Neurosci 2005; 25: 11489–11493.
Kanat M, Heinrichs M, Schwarzwald R, Domes G . Oxytocin attenuates neural reactivity to masked threat cues from the eyes. Neuropsychopharmacology 2015; 40: 287–295.
Andari E, Duhamel JR, Zalla T, Herbrecht E, Leboyer M, Sirigu A . Promoting social behavior with oxytocin in high-functioning autism spectrum disorders. Proc Natl Acad Sci USA 2010; 107: 4389–4394.
Bakermans-Kranenburg MJ, van IJMH . Sniffing around oxytocin: review and meta-analyses of trials in healthy and clinical groups with implications for pharmacotherapy. Transl Psychiatry 2013; 3: e258.
Bartz JA, Zaki J, Bolger N, Hollander E, Ludwig NN, Kolevzon A et al. Oxytocin selectively improves empathic accuracy. Psychol Sci 2010; 21: 1426–1428.
Guastella AJ, Einfeld SL, Gray KM, Rinehart NJ, Tonge BJ, Lambert TJ et al. Intranasal oxytocin improves emotion recognition for youth with autism spectrum disorders. Biol Psychiatry 2010; 67: 692–694.
Domes G, Heinrichs M, Kumbier E, Grossmann A, Hauenstein K, Herpertz SC . Effects of intranasal oxytocin on the neural basis of face processing in autism spectrum disorder. Biol Psychiatry 2013; 74: 164–171.
Guastella AJ, Howard AL, Dadds MR, Mitchell P, Carson DS . A randomized controlled trial of intranasal oxytocin as an adjunct to exposure therapy for social anxiety disorder. Psychoneuroendocrinology 2009; 34: 917–923.
Labuschagne I, Phan KL, Wood A, Angstadt M, Chua P, Heinrichs M et al. Oxytocin attenuates amygdala reactivity to fear in generalized social anxiety disorder. Neuropsychopharmacology 2010; 35: 2403–2413.
Bertsch K, Gamer M, Schmidt B, Schmidinger I, Walther S, Kastel T et al. Oxytocin and reduction of social threat hypersensitivity in women with borderline personality disorder. Am J Psychiatry 2013; 170: 1169–1177.
Kumsta R, Heinrichs M . Oxytocin, stress and social behavior: neurogenetics of the human oxytocin system. Curr Opin Neurobiol 2013; 23: 11–16.
Ebstein RP, Israel S, Chew SH, Zhong S, Knafo A . Genetics of human social behavior. Neuron 2010; 65: 831–844.
Jacob S, Brune C, Carter C, Leventhal B, Lord C, Cookjr E . Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neurosci Lett 2007; 417: 6–9.
Lerer E, Levi S, Salomon S, Darvasi A, Yirmiya N, Ebstein RP . Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Mol Psychiatry 2008; 13: 980–988.
Liu X, Kawamura Y, Shimada T, Otowa T, Koishi S, Sugiyama T et al. Association of the oxytocin receptor (OXTR) gene polymorphisms with autism spectrum disorder (ASD) in the Japanese population. J Hum Genet 2010; 55: 137–141.
Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M et al. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biol Psychiatry 2005; 58: 74–77.
Chen FS, Kumsta R, von Dawans B, Monakhov M, Ebstein RP, Heinrichs M . Common oxytocin receptor gene (OXTR) polymorphism and social support interact to reduce stress in humans. Proc Natl Acad Sci USA 2011; 108: 19937–19942.
Kim HS, Sherman DK, Sasaki JY, Xu J, Chu TQ, Ryu C et al. Culture, distress, and oxytocin receptor polymorphism (OXTR) interact to influence emotional support seeking. Proc Natl Acad Sci USA 2010; 107: 15717–15721.
Kogan A, Saslow LR, Impett EA, Oveis C, Keltner D, Rodrigues Saturn S . Thin-slicing study of the oxytocin receptor (OXTR) gene and the evaluation and expression of the prosocial disposition. Proc Natl Acad Sci USA 2011; 108: 19189–19192.
Rodrigues SM, Saslow LR, Garcia N, John OP, Keltner D . Oxytocin receptor genetic variation relates to empathy and stress reactivity in humans. Proc Natl Acad Sci USA 2009; 106: 21437–21441.
Krueger F, Parasuraman R, Iyengar V, Thornburg M, Weel J, Lin M et al. Oxytocin receptor genetic variation promotes human trust behavior. Front Hum Neurosci 2012; 6: 4.
Bakermans-Kranenburg MJ, van Ijzendoorn MH . Oxytocin receptor (OXTR) and serotonin transporter (5-HTT) genes associated with observed parenting. Soc Cogn Affect Neurosci 2008; 3: 128–134.
Inoue H, Yamasue H, Tochigi M, Abe O, Liu X, Kawamura Y et al. Association between the oxytocin receptor gene and amygdalar volume in healthy adults. Biol Psychiatry 2010; 68: 1066–1072.
Tost H, Kolachana B, Hakimi S, Lemaitre H, Verchinski BA, Mattay VS et al. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proc Natl Acad Sci USA 2010; 107: 13936–13941.
Tost H, Kolachana B, Verchinski BA, Bilek E, Goldman AL, Mattay VS et al. Neurogenetic effects of OXTR rs2254298 in the extended limbic system of healthy Caucasian adults. Biol Psychiatry 2011; 70: e37–e39, author reply e41-32.
Furman DJ, Chen MC, Gotlib IH . Variant in oxytocin receptor gene is associated with amygdala volume. Psychoneuroendocrinology 2011; 36: 891–897.
Lischke A, Berger C, Prehn K, Heinrichs M, Herpertz SC, Domes G . Intranasal oxytocin enhances emotion recognition from dynamic facial expressions and leaves eye-gaze unaffected. Psychoneuroendocrinology 2012; 37: 475–481.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B et al. The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229.
Binder A . Identification of genes for a complex trait: examples from hypertension. Curr Pharm Biotechnol 2006; 7: 1–13.
Binder A, Garcia E, Wallace C, Gbenga K, Ben-Shlomo Y, Yarnell J et al. Haplotypes of the beta-2 adrenergic receptor associate with high diastolic blood pressure in the Caerphilly prospective study. J Hypertens 2006; 24: 471–477.
Bryant RA, Hung L, Dobson-Stone C, Schofield PR . The association between the oxytocin receptor gene (OXTR) and hypnotizability. Psychoneuroendocrinology 2013; 38: 1979–1984.
Moons WG, Way BM, Taylor SE . Oxytocin and vasopressin receptor polymorphisms interact with circulating neuropeptides to predict human emotional reactions to stress. Emotion 2014; 14: 562–572.
Peltola MJ, Yrttiaho S, Puura K, Proverbio AM, Mononen N, Lehtimaki T et al. Motherhood and oxytocin receptor genetic variation are associated with selective changes in electrocortical responses to infant facial expressions. Emotion 2014; 14: 469–477.
Schneiderman I, Kanat-Maymon Y, Ebstein RP, Feldman R . Cumulative risk on the oxytocin receptor gene (OXTR) underpins empathic communication difficulties at the first stages of romantic love. Soc Cogn Affect Neurosci 2014; 9: 1524–1529.
Tabak BA, McCullough ME, Carver CS, Pedersen EJ, Cuccaro ML . Variation in oxytocin receptor gene (OXTR) polymorphisms is associated with emotional and behavioral reactions to betrayal. Soc Cogn Affect Neurosci 2014; 9: 810–816.
Steyer R, Schwenkmezger P, Notz P, Eid M . Der Mehrdimensionale Befindlichkeitsfragebogen (MDBF). Hogrefe: Göttingen, 1997.
Montag C, Fiebach CJ, Kirsch P, Reuter M . Interaction of 5-HTTLPR and a variation on the oxytocin receptor gene influences negative emotionality. Biol Psychiatry 2011; 69: 601–603.
Dudbridge F . Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered 2008; 66: 87–98.
Walter NT, Montag C, Markett S, Felten A, Voigt G, Reuter M . Ignorance is no excuse: moral judgments are influenced by a genetic variation on the oxytocin receptor gene. Brain Cogn 2012; 78: 268–273.
Wade M, Hoffmann TJ, Wigg K, Jenkins JM . Association between the oxytocin receptor (OXTR) gene and children's social cognition at 18 months. Genes Brain Behav 2014; 13: 603–610.
Marsh AA, Kozak MN, Ambady N . Accurate identification of fear facial expressions predicts prosocial behavior. Emotion 2007; 7: 239–251.
Uljarevic M, Hamilton A . Recognition of emotions in autism: a formal meta-analysis. J Autism Dev Disord 2013; 43: 1517–1526.
Domes G, Schulze L, Herpertz SC . Emotion recognition in borderline personality disorder-a review of the literature. J Personal Disord 2009; 23: 6–19.
Averbeck BB, Bobin T, Evans S, Shergill SS . Emotion recognition and oxytocin in patients with schizophrenia. Psychol Med 2012; 42: 259–266.
Chen FS, Johnson SC . An oxytocin receptor gene variant predicts attachment anxiety in females and autism-spectrum traits in males. Soc Psychol Personal Sci 2012; 3: 93–99.
Domes G, Lischke A, Berger C, Grossmann A, Hauenstein K, Heinrichs M et al. Effects of intranasal oxytocin on emotional face processing in women. Psychoneuroendocrinology 2010; 35: 83–93.
Kumsta R, Hummel E, Chen FS, Heinrichs M . Epigenetic regulation of the oxytocin receptor gene: implications for behavioral neuroscience. Front Neurosci 2013; 7: 83.
Kusui C, Kimura T, Ogita K, Nakamura H, Matsumura Y, Koyama M et al. DNA methylation of the human oxytocin receptor gene promoter regulates tissue-specific gene suppression. Biochem Biophys Res Commun 2001; 289: 681–686.
Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med 2009; 7: 62.
Dadds MR, Moul C, Cauchi A, Dobson-Stone C, Hawes DJ, Brennan J et al. Methylation of the oxytocin receptor gene and oxytocin blood levels in the development of psychopathy. Dev Psychopathol 2014; 26: 33–40.
Lerer E, Levi S, Israel S, Yaari M, Nemanov L, Mankuta D et al. Low CD38 expression in lymphoblastoid cells and haplotypes are both associated with autism in a family-based study. Autism Res 2010; 3: 293–302.
Sauer C, Montag C, Worner C, Kirsch P, Reuter M . Effects of a common variant in the CD38 gene on social processing in an oxytocin challenge study: possible links to autism. Neuropsychopharmacology 2012; 37: 1474–1482.
Heinrichs M, Domes G . Neuropeptides and social behaviour: effects of oxytocin and vasopressin in humans. Prog Brain Res 2008; 170: 337–350.
Heinrichs M, Chen FS, Domes G. Oxytocin. In: Hofman S (ed). Psychobiological Approaches for Anxiety Disorders: Treatment Combination Strategies. Wiley-Blackwell: Oxford, 2012, pp 123–143.
McMahon FJ, Insel TR . Pharmacogenomics and personalized medicine in neuropsychiatry. Neuron 2012; 74: 773–776.
Ozomaro U, Wahlestedt C, Nemeroff CB . Personalized medicine in psychiatry: problems and promises. BMC Med 2013; 11: 132.
Acknowledgements
We thank Dr Valentina Colonnello for her support in creating study materials; Dr Gail Alvarez, Celia Gaertig, Valentina Glueck, Laura Huber, Anna Mauz and Richard Rau for their assistance during data collection; and Hanne Collins for editorial assistance. This study was supported by Deutsche Forschungsgemeinschaft research grant HE 5310/1-1 to MH, FSC and RK. RK gratefully acknowledges grant support from the Deutsche Forschungsgemeinschaft (DFG; KU 2479/3-1). GD and MH gratefully acknowledge grant support from the DFG (Do1312/2-1; DFG, Do1312/2-3). MH was supported by the European Neuroscience Network NEUREX. FSC gratefully acknowledges support from a research fellowship of the Alexander von Humboldt Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, F., Kumsta, R., Dvorak, F. et al. Genetic modulation of oxytocin sensitivity: a pharmacogenetic approach. Transl Psychiatry 5, e664 (2015). https://doi.org/10.1038/tp.2015.163
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2015.163
This article is cited by
-
Randomized clinical trial shows no substantial modulation of empathy-related neural activation by intranasal oxytocin in autism
Scientific Reports (2021)
-
The Neurogenetics of Racial Injustice: Oxytocin Receptor (OXTR) Gene rs53576 is ASSOCIATED with Perceived Discrimination and Other-Oriented Justice Beliefs in African Americans
Race and Social Problems (2021)
-
Oxytocin bei Schizophrenie
Der Nervenarzt (2019)
-
Genetic Variation in the Maternal Oxytocin System Affects Cortisol Responsiveness to Breastfeeding in Infants and Mothers
Adaptive Human Behavior and Physiology (2018)
-
Additive effects of oxytocin receptor gene polymorphisms on reward circuitry in youth with autism
Molecular Psychiatry (2017)