Abstract
We have used a translational Convergent Functional Genomics (CFG) approach to discover genes involved in alcoholism, by gene-level integration of genome-wide association study (GWAS) data from a German alcohol dependence cohort with other genetic and gene expression data, from human and animal model studies, similar to our previous work in bipolar disorder and schizophrenia. A panel of all the nominally significant P-value SNPs in the top candidate genes discovered by CFG (n=135 genes, 713 SNPs) was used to generate a genetic risk prediction score (GRPS), which showed a trend towards significance (P=0.053) in separating alcohol dependent individuals from controls in an independent German test cohort. We then validated and prioritized our top findings from this discovery work, and subsequently tested them in three independent cohorts, from two continents. In order to validate and prioritize the key genes that drive behavior without some of the pleiotropic environmental confounds present in humans, we used a stress-reactive animal model of alcoholism developed by our group, the D-box binding protein (DBP) knockout mouse, consistent with the surfeit of stress theory of addiction proposed by Koob and colleagues. A much smaller panel (n=11 genes, 66 SNPs) of the top CFG-discovered genes for alcoholism, cross-validated and prioritized by this stress-reactive animal model showed better predictive ability in the independent German test cohort (P=0.041). The top CFG scoring gene for alcoholism from the initial discovery step, synuclein alpha (SNCA) remained the top gene after the stress-reactive animal model cross-validation. We also tested this small panel of genes in two other independent test cohorts from the United States, one with alcohol dependence (P=0.00012) and one with alcohol abuse (a less severe form of alcoholism; P=0.0094). SNCA by itself was able to separate alcoholics from controls in the alcohol-dependent cohort (P=0.000013) and the alcohol abuse cohort (P=0.023). So did eight other genes from the panel of 11 genes taken individually, albeit to a lesser extent and/or less broadly across cohorts. SNCA, GRM3 and MBP survived strict Bonferroni correction for multiple comparisons. Taken together, these results suggest that our stress-reactive DBP animal model helped to validate and prioritize from the CFG-discovered genes some of the key behaviorally relevant genes for alcoholism. These genes fall into a series of biological pathways involved in signal transduction, transmission of nerve impulse (including myelination) and cocaine addiction. Overall, our work provides leads towards a better understanding of illness, diagnostics and therapeutics, including treatment with omega-3 fatty acids. We also examined the overlap between the top candidate genes for alcoholism from this work and the top candidate genes for bipolar disorder, schizophrenia, anxiety from previous CFG analyses conducted by us, as well as cross-tested genetic risk predictions. This revealed the significant genetic overlap with other major psychiatric disorder domains, providing a basis for comorbidity and dual diagnosis, and placing alcohol use in the broader context of modulating the mental landscape.
Similar content being viewed by others
Introduction
‘Drunkenness is nothing but voluntary madness.’—Seneca
Alcohol use and overuse (alcoholism) havedeep historical and cultural roots, as well as important medical and societal consequences.1 Whereas there is evidence for roles for both genes and environment in alcoholism, a comprehensive biological understanding of the disorder has been elusive so far, despite extensive work in the field. Most notably, there has been until recently insufficient translational integration across functional and genetic studies, and across human and animal model studies, resulting in missed opportunities for a comprehensive understanding.
As part of a translational Convergent Functional Genomics (CFG) approach, developed by us over the last 15 years,2 and expanding upon our earlier work on identifying genes for alcoholism,3, 4, 5 we set out to comprehensively identify candidate genes, pathways and mechanisms for alcoholism, integrating the available evidence in the field to date. We have used data from a published German genome-wide association study for alcoholism.6 We integrated those data in a Bayesian-like manner with other human genetic data (association or linkage) for alcoholism, as well as human gene expression data, post-mortem brain gene expression data and peripheral (blood and cell culture) gene expression data. We also used relevant animal model genetic data (transgenic and quantitative trait loci (QTL)), as well as animal model gene expression data (brain and blood) generated by our group andothers (Figures 1 and 2). Human data provide specificity for the illness, and animal model data provide sensitivity of detection. Together, they helped to identify and prioritize candidate genes for the illness using a polyevidence CFG score, resulting in essence in a de facto field-wide integration putting together all the available lines of evidence to date. Once that is done, biological pathway analyses can be conducted and mechanistic models can be constructed.
An obvious next step is developing a way of applying that knowledge to genetic testing of individuals to determine risk for the disorder. On the basis of our comprehensive identification of top candidate genes described in this paper, we have chosen all the nominally significant P-value SNPs corresponding to each of those 135 genes from the GWAS data set used for discovery (top candidate genes prioritized by CFG with the score of 8 and above (≥50% maximum possible CFG score of 16) and assembled a Genetic Risk Prediction panel out of those 713 SNPs. We then developed a Genetic Risk Prediction Score (GRPS) for alcoholism based on the presence or absence of the alleles of the SNPs associated with the illness from the discovery GWAS, and tested the GRPS in an independent German cohort,51 to see whether it can differentiate alcohol-dependent subjects from controls, observing a trend towards significance.
In order to validate and prioritize genes in this panel using a behavioral prism, we then looked at the overlap between our panel of 135 top candidate genes and genes changed in expression in a stress-reactive animal model for alcoholism developed by our group, the DBP knockout mouse.4,5 We used this overlap to reduce our panel to 11 genes (66 SNPs).
This small panel of 11 genes was subsequently tested and shown to be able to differentiate between alcoholics and controls in the three independent test cohorts, one German51 and two US-based,52 suggesting that the animal model served in essence as a filter to identify from the larger list of CFG-prioritized genes the key behaviorally relevant genes. Our results indicate that panels of SNPs in top genes identified and prioritized by CFG analysis and by a behaviorally relevant animal model can differentiate between alcoholics and controls at a population level (Figure 3), although at an individual level the margin may be small (Supplementary Figure S2). The latter point suggests that, similar to bipolar disorder53 and schizophrenia,54 the contextual cumulative combinatorics of common gene variants and environment55 has a major role in risk for illness.
Lastly, we have looked at overlap with genes for other major psychiatric disorder domains (bipolar disorders, anxiety disorders, schizophrenias) from our previous studies and provide evidence for shared genes (Figures 4 and 5) as well as shared genetic risk (Figure 6).
Genetic load for bipolar disorder and schizophrenia in alcoholism. A total of 34 out of 66 SNPs in our alcohol GRPS-11 panel (current work; in n=10 genes), 42 out of 224 SNPs in our bipolar GRPS53 (in n=34 genes) and 151 out of 542 SNPs in our schizophrenia GRPS54 (in n=35 genes) were present and tested in the alcohol cohorts 3 and 4. See also Supplementary Table S7.
Overall, this work sheds light on the genetic architecture and pathophysiology of alcoholism, provides mechanistic targets for therapeutic intervention and has implications for genetic testing to assess risk for illness before the illness manifests itself clinically, opening the door for enhanced prevention strategies at a young age. As alcoholism is a disease that does not exist if the exogenous agent (alcohol) is not consumed, the use of genetic information to inform lifestyle choices could be quite powerful.
Materials and methods
Human subject cohorts
Discovery cohort (cohort 1): GWAS for alcohol dependence from Germany
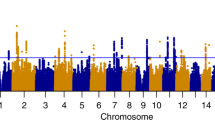
Data for the discovery CFG work (Cohort 1) were obtained from a GWA study of self-reported German descent subjects, consisting of 411 alcohol-dependent male subjects and 1307 population-based controls (663 male and 644 female subjects).6 Individuals were genotyped using HumanHap 550 BeadChips (Illumina Inc, San Diego, CA, USA). SNPs with a nominal allelic P-value <0.05 were selected for analysis. No Bonferroni correction was performed.
Test cohort 2 (alcohol dependence, Germany)
An independent test cohort of German descent51 consisting of 740 alcohol-dependent male subjects and 861 controls (276 male and 585 female subjects) was used for testing the results of the discovery analyses. Individuals were genotyped using Illumina Human610Quad or Illumina Human660w Quad BeadChips (Illumina Inc). The controls were genotyped using Illumina HumanHap550 Bead Chips.
Test cohort 3 (alcohol dependence, United States) and test cohort 4 (alcohol abuse, United States)
The sample consisted of small nuclear families originally collected for linkage studies, and unrelated individuals, Caucasians and African-American, male and female subjects. The subjects were recruited at five US clinical sites: Yale University School of Medicine (APT Foundation; New Haven, CT, USA), the University of Connecticut Health Center (Farmington, CT, USA), the University of Pennsylvania Perelman School of Medicine (Philadelphia, PA, USA), the Medical University of South Carolina (Charleston, SC, USA) and McLean Hospital (Belmont, MA, USA). All subjects were interviewed using the Semi-Structured Assessment for Drug Dependence and Alcoholism to derive diagnoses for lifetime alcohol dependence, alcohol abuse and other major psychiatric traits according to the DSM-IV criteria. There were 1687 male subjects with alcohol dependence, 366 male subjects with alcohol abuse and 475 male controls. There were 1081 female subjects with alcohol dependence, 234 female subjects with alcohol abuse and 786 female controls (Table 1). Individuals were genotyped on the Illumina HumanOmni1-Quad v1.0 microarray (988 306 autosomal SNPs). GWAS genotyping was conducted at the Yale Center for Genome Analysis and the Center for Inherited Disease Research. Genotypes were called using the GenomeStudio software V2011.1 and genotyping module version 1.8.4 (Illumina Inc).52
Gene identification in discovery cohort 1
Quality control
Genotype data had been filtered using stringent quality-control criteria as described earlier51 and accounted for call rate, population substructure, cryptic relatedness, minor allele frequency and batch effects.
Association test in discovery sample
Association testing was performed using PLINK 1.07 (http://pngu.mgh.harvard.edu/~purcell)56 software package. A logistic regression modelling approach was applied to correct for population stratification. Therefore, principal component analysis was conducted considering only independent autosomal SNPs with minor allele frequency >0.05 and pairwise R2<0.05 within a 200-SNP window. LD filtering resulted in a set of 28 505 SNPs used for principal component analysis, which was carried out using GCTA 1.04 (http://www.complextraitgenomics.com/software/gcta/).57 The first two principal components resulting from this analysis were included as covariates in the logistic regression model.
Assignment of SNPs to genes
Genes corresponding to SNPs were identified initially using the annotation file from the Illumina website (http://www.illumina.com, HumanHAP550v3_Gene_Annotation). Next, genes were cross-checked with GeneCards (http://www.genecards.org) to ensure that each gene symbol was current. Any gene symbol that matched to a different gene symbol in Gene Cards was checked to verify chromosome number and location match with the original gene, and was replaced with the current GeneCards gene symbol. SNPs from the original annotation files that had no gene matches in the annotation file and UCSC Genome Browser (that is, not falling within an exon or intron of a known gene) were assumed to regulate and thus implicate the gene closest to the SNP location, using the refSNP database from NCBI (http://www.ncbi.nlm.nih.gov/snp/?SITE=NcbiHome&submit=Go).
Convergent functional genomic analyses
Databases
We have established in our laboratory (Laboratory of Neurophenomics, Indiana University School of Medicine, www.neurophenomics.info) manually curated databases of all the human gene expression (post-mortem brain, blood and cell cultures), human genetic (association, copy number variants (CNVs) and linkage), animal model genetic and animal model gene expression studies published to date on psychiatric disorders. Only the findings deemed significant in the primary publication, by the study authors, using their particular experimental design and thresholds, are included in our databases. Our databases include only primary literature data and do not include review papers or other secondary data integration analyses to avoid redundancy and circularity. These large and constantly updated databases have been used in our CFG cross-validation and prioritization (Figure 1).
Human post-mortem brain, blood and other peripheral tissue gene expression evidence
Information about genes was obtained and imported in our databases searching the primary literature with PubMed (http://ncbi.nlm.nih.gov/PubMed), using various combinations of keywords. For this work, the keywords were as follows: alcohol, alcoholism, human, brain, postmortem, lymphocytes, blood, cells and gene expression.
Human genetic evidence (association, linkage)
To designate convergence for a particular gene, the gene had to have independent published evidence of association or linkage for alcoholism. We sought to avoid using any association studies that included subjects who were also included in our discovery or test cohorts. For linkage, the location of each gene was obtained through GeneCards (http://www.genecards.org), and the sex-averaged cM location of the start of the gene was then obtained through http://compgen.rutgers.edu/old/map-interpolator/. For linkage convergence, per our previously published criteria, the start of the gene had to map within 5 cM of the location of a marker linked to the disorder with a lod score of ≥2.
Animal model brain and blood gene expression evidence
For animal model brain and blood gene expression evidence, we have used our own rat model data sets,3 as well as published reports from the literature curated in our databases.
The rat animal model experimental work from our group was previously described.3 The experimental approaches used to produce the animal model data for CFG analysis were carried out in two rat lines selectively bred for divergent alcohol preference: inbred alcohol-preferring (iP) versus inbred alcohol-non-preferring (iNP) rats. Following five brain regions were chosen for gene expression studies in these rat lines: the frontal cortex, amygdala, caudate–putamen, nucleus accumbens and hippocampus. Animal studies, as well as human imaging and post-mortem analyses, had previously provided evidence that these regions are implicated in alcoholism.
Data for the analysis came from studies of three experimental paradigms. Paradigm 1 examined basal level of gene expression in the brains of the alcohol-naive iP and iNP lines of rats. This basal comparison was performed to determine innate differences between these two lines with a marked divergence in the willingness to consume alcohol. We hypothesized that the innate differences in gene expression between the iP and iNP would involve some of the genes associated with an increased susceptibility for alcohol dependence. Paradigm 2 examined the effects of chronic 24-h free-choice alcohol consumption on gene expression in iP rats compared with alcohol-naive iP rats. This paradigm looked for gene expression changes in the brain associated with the direct influence of peripherally self-administered alcohol in the genetically susceptible rats. In Paradigm 3, iP rats were allowed to self-infuse alcohol directly into the posterior ventral tegmental area, the originating area of the mesolimbic dopamine system. The advantage of this latter procedure is that it isolates the neurocircuitry involved in alcohol reinforcement, and eliminates the peripheral effects of alcohol. Following the establishment of alcohol self-administration into the posterior VTA, gene expression levels in target brain areas were measured and compared with P rats that received artificial cerebral spinal fluid infusions into the posterior VTA.
Animal model genetic evidence
To search for mouse genetic evidence (transgenic and QTL) for our candidate genes, we utilized PubMed as well as the Mouse Genome Informatics (http://www.informatics.jax.org; Jackson Laboratory, Bar Harbor, ME, USA) database, and used the search ‘Genes and Markers’ form to find transgenic in categories for abnormal alcohol consumption, alcohol preference, alcohol aversion, impaired behavioral response to alcohol, hyperactivity elicited by ethanol administration and enhanced behavioral response to alcohol. For QTL convergence, the start of the gene had to map within 5 cM of the location of these markers.
CFG scoring
We used a nominal P-value threshold (having at least one SNP with P<0.05) for including genes from the discovery GWAS in the CFG analysis. No Bonferroni correction was performed.
Internal score: For each of these genes implicated by SNPs, we calculated the percent of SNPs that were nominally significant (ratio of number of nominally significant SNPs over total number of SNPs tested for that gene, multiplied by 100), obtaining a distribution of values. The genes in the top 0.1% of the distribution were given an internal score of 4 points, those in the top 5% of the distribution were given 3 points and the remaining genes all received 2 points. The internal score provides a prioritization of genes based on GWAS results and might prioritize genes that have higher biological relevance and heterogeneity.
External score: Human and animal model data, genetic and gene expression were integrated and tabulated, resulting in a polyevidence CFG score. All six cross-validating lines of evidence (human data and animal model data) were weighted such that evidence from human studies was prioritized 2x over evidence from animal models, gene expression evidence was prioritized 2x over genetic evidence and brain evidence was prioritized 2x over peripheral tissue evidence (Figure 1). For human genetic evidence, 2 points were assigned if it was from association and 1 point if it was from linkage studies. For animal model genetic evidence, 2 points if it was from transgenic and 1 point if it was from QTL. The maximum possible external score for each gene is 12.
We have capped (one positive study scores maximum points) the hypothesis-driven candidate gene genetic association evidence and animal model genetic (transgenic) lines of evidence, regardless of how many other such studies support that gene, to avoid potential ‘popularity’ biases, where some genes are more studied than others. For discovery-driven gene expression studies, we have capped (one positive study scores maximum points) the human post-mortem brain work because of the paucity of brain collections and the fact that such studies often use the same brain bank sources. However, we have not similarly capped the animal model brain and blood gene expression evidence, as such studies are not only discovery-based, but use independent cohorts of animals. These were scored differentially, based on the number of studies showing evidence for a given gene: three or more different studies received full maximum points, two studies 0.75 of maximum points and one study 0.5 of the maximum points. Our group generated data sets for three independent animal studies for this analysis (see above).
The more lines of evidence for a gene—that is, the more times a gene shows up as a positive finding across independent studies, platforms, methodologies and species—the higher its external CFG score (Figure 1). This is similar conceptually to the Google PageRank algorithm, in which the more links to a page, the higher it comes up on the search prioritization list. It has not escaped our attention that other ways of weighing the lines of evidence may give slightly different results in terms of prioritization, if not in terms of the list of genes per se. Nevertheless, we think this simple scoring system provides a good separation of genes, with specificity provided by human data and sensitivity provided by animal model data.
Prioritizing top alcoholism candidate genes that overlap with a stress-reactive animal model of alcoholism
Stress has been proposed as a driver of alcoholism, notably by Koob and colleagues,58,59 as well as by Heilig and colleagues.60 We have previously identified the circadian clock gene DBP as a candidate gene for bipolar disorder,61 as well as for alcoholism,3 using a CFG approach. In follow-up work, we established mice with a homozygous deletion of DBP (DBP KO) as a stress-reactive genetic animal model of bipolar disorder and alcoholism.4 We reported that DBP KO mice have lower locomotor activity, blunted responses to stimulants and gain less weight over time. In response to a stress paradigm that translationally mimics what can happen in humans (chronic stress-isolation housing for 4 weeks, with acute stress, on top of that- experimental handling in week 3), the mice exhibit a diametric switch in these phenotypes. DBP KO mice are also activated by sleep deprivation, similar to bipolar patients, and that activation is prevented by treatment with the mood stabilizer drug valproate. Moreover, these mice show increased alcohol intake following exposure to stress. Microarray studies of brain and blood revealed a pattern of gene expression changes that may explain the observed phenotypes. CFG analysis of the gene expression changes identified a series of candidate genes and blood biomarkers for bipolar disorder, alcoholism and stress reactivity. Subsequent studies by us showed that treatment with the omega-3 fatty acid docosahexaenoic acid (DHA) normalized the gene expression (brain and blood) and behavioral phenotypes of this mouse model, including reducing alcohol consumption.5
We examined the overlap between the top candidate genes for alcoholism from the current analysis and the top candidate genes from the DBP KO stress mice, thus reducing the list from 135 to 11 (Figure 4).
Pathway analyses
IPA 9.0 (Ingenuity Systems, www.ingenuity.com, Redwood City, CA, USA) was used to analyze the biological roles, including top canonical pathways and diseases, of the candidate genes resulting from our work (Table 2 and Supplementary Table S2), as well as used to identify genes in our data sets that are the targets of existing drugs (Supplementary Table S3). Pathways were identified from the IPA library of canonical pathways that were most significantly associated with genes in our data set. The significance of the association between the data set and the canonical pathway was measured in 2 ways: (1) a ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed; (2) Fisher’s exact test was used to calculate a P-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone. We also conducted a KEGG pathway analysis through the Partek Genomic Suites 6.6 software package, Partek Inc, Saint Louis, MO, USA), and GeneGo MetaCore from Thomson Reuters, New York, NY, USA) pathway analyses (https://portal.genego.com/).
Epistasis testing
The test cohort 2 data were used to test for epistatic interactions among the best P-value SNPs in the 11 top candidate genes from our work. SNP–SNP allelic epistasis was tested for each distinct pair of SNPs between genes, using the PLINK software package (Supplementary Table S5).
Genetic risk prediction
The software package PLINK 1.07 (http://pngu.mgh.harvard.edu/~purcell)56 was used to extract individual genotype information for each subject from the test cohorts 2, 3 and 4 data files.
As we had previously performed for bipolar disorder and schizophrenia, we developed a polygenic GRPS for alcoholism based on the presence or absence of the alleles of the SNPs associated with illness in the discovery GWAS cohort 1, and tested the GRPS in three independent cohorts, from different geographic areas, ethnicities and different types of alcoholism. We tested two panels: a larger panel containing all the nominally significant SNPs in top CFG scoring candidate genes (n=135) from the discovery GWAS1 in the top CFG-prioritized genes (Supplementary Tables S1 and S4) and a smaller one (n=11) containing genes out of the larger panel that were cross-validated using an animal model of alcoholism.
Of note, our genes, SNP panels and choice of affected alleles were based solely on analysis of the discovery GWAS1, which is our discovery cohort, completely independently from the test cohorts. Each SNP has two alleles (represented by base letters at that position). One of them is associated with the illness (affected), the other not (non-affected), based on the odds ratios from the discovery GWAS1. We assigned the affected allele a score of 1 and the non-affected allele a score of 0. A two-dimensional matrix of subjects by GRP panel alleles is generated, with the cells populated by 0 or 1. A SNP in a particular individual subject can have any permutation of 1 and 0 (1 and 1, 0 and 1, 0 and 0). By adding these numbers, the minimum score for a SNP in an individual subject is 0, and the maximum score is 2. By adding the scores for all the alleles in the panel, averaging that and multiplying by 100, we generated for each subject an average score corresponding to a genetic loading for disease, which we call Genetic Risk Predictive Score.53,54
To test for significance, a one-tailed t-test with unequal variance was performed between the alcoholic subjects and the control subjects, looking at differences in GRPS.
Receiver operating characteristic curves
Receiver operating characteristic curves were plotted using IBM SPSS Statistics 21. Diagnosis was converted to a binary call of 0 (control) or 1 (alcohol-dependent or abuser) and entered as the state variable, with calculated GRPS entered as the test variable (Supplementary Figure S2).
Results
Top candidate genes
To minimize false-negatives, we initially cast a wide net, using as a filter a minimal requirement for a gene to have both some GWAS evidence and some additional independent evidence. Thus, out of the 6085 genes with at least a SNP at P<0.05 in the discovery GWAS cohort 1, we generated a list of 3142 genes that also had some additional line of evidence (human or animal model data), implicating them in alcoholism (CFG score ≥2.5 (≥2 internal)+(≥0.5 external)). This suggests, using these minimal thresholds and requirements, that the repertoire of genes potentially involved directly or indirectly in alcohol consumption and alcoholism may be quite large, similar to what we have previously seen for bipolar disorder62 and schizophrenia.54 To minimize false-positives, we used an internal score based on percent of SNPs in a gene that were nominally significant, with 4 points for those in the top 0.1% of the distribution (n=77), 3 points for those in the top 5% of the distribution (n=561) and 2 points for the rest of the nominally significant SNPs (n=5447). We then used the CFG analysis and scoring integrating multiple lines of evidence to prioritize this list of genes (Figure 1) and focused our subsequent analyses on only the top CFG scoring candidate genes. Overall, 135 genes had a CFG score of 8 and above (≥50% of maximum possible score of 16).
Of note, there was no correlation between CFG prioritization and gene size, thus excluding a gene-size effect for the observed enrichment (Supplementary Figure S1).
Biological pathways and drug targets
Pathway analyses were carried out on the top candidate genes (Table 2). Notably, Gαi signaling, cocaine addiction and transmission of nerve impulses were the top biological pathways in alcoholism, which may be informative for treatments and drug discovery efforts by pharmaceutical companies. Of note, these top candidate genes were identified and prioritized only for evidence for alcoholism before pathway analyses; therefore, the overlap with cocaine addiction is a completely independent result, suggesting a shared drive and neurobiology. Consistent with that, two of our 135 top candidate genes for alcoholism (CPE and VWF) had SNPs with P<10−5 in a recent GWAS of cocaine addiction.63
Some of the top alcohol candidate genes have prior evidence of being modulated by the omega-3 fatty acid DHA in our DBP mouse animal model (Table 3 and Supplementary Table S1). That is of particular interest, as we have previously shown that treatment with the omega-3 fatty acid DHA decreased alcohol consumption in that animal model, as well as in another independent animal model, the alcohol-preferring P rats.5 Omega-3 fatty acids, particularly DHA, have been described to have alcoholism, mood, psychosis and suicide-modulating properties, in preclinical models as well as some human clinical trials and epidemiological studies. For example, deficits in omega-3 fatty acids have been linked to increased depression and aggression in animal models64,65 and humans.66,67 DHA prevents ethanol damage in vitro in rat hippocampal slices.68 Omega-3 supplementation can prevent oxidative damage caused by prenatal alcohol exposure in rats.69 Of note, deficits in DHA have been reported in erythrocytes70 and in the post-mortem orbitofrontal cortex of patients with bipolar disorder, and were greater in those that had high versus those that had low alcohol abuse.71 Low DHA levels may be a risk factor for suicide.72,73 Omega-3 fatty acids have been reported to be clinically useful in the treatment of both mood74, 75, 76, 77 and psychotic disorders.78, 79, 80
Other existing pharmacological drugs that modulate alcohol candidate genes identified by us include, besides benzodiazepines, dopaminergic agents, glutamatergic agents, serotonergic agents, as well as statins (Supplementary Table S3).
Genetic risk prediction score
Once the genes involved in a disorder are identified, and prioritized for likelihood of involvement, then an obvious next step is developing a way of applying that knowledge to genetic testing of individuals to determine risk for the disorder. On the basis of our identification of top candidate genes described above using CFG, we pursued a polygenic panel approach, with digitized binary scoring for presence or absence, similar to the one we have devised and used in the past for biomarkers testing53,81 and for genetic testing in bipolar disorder53 and schizophrenia.54 Somewhat similar approaches but without CFG prioritization, attempted by other groups, have been either unsuccessful82 or have required very large panels of markers.83
We chose all the nominally significant P-value SNPs (P<0.05) in each of our top CFG-prioritized genes (n=135 with CFG score ≥8; Supplementary Table S1) in the GWAS1 data set used for discovery, and assembled a GRPS-135 panel out of those SNPs (Table 4). We then tested the GRPS-135 in the independent German test cohort 2, based on the presence or absence of the alleles of the SNPs associated with the illness, comparing the alcoholic subjects to controls (Table 4), and showed that, although there was a trend, we were not able to distinguish alcoholics from controls in both independent test cohorts.
We then prioritized a smaller panel of 11 genes (Table 3) out of this larger panel, by using as a cross-validator the top genes from a stress-reactive mouse animal model for alcoholism, the DBP knockout mouse4 (Figure 4). The small panel (GRPS-11) showed more robust results than the larger panel (Table 4), suggesting that it captures the key behaviorally relevant genes.
Discussion
Our CFG approach helped to prioritize a very rich-in-signal and biologically interesting set of genes (Table 3 and Supplementary Table S1). Some, such as SNCA, CPE, DRD2 and GRM3, have weaker evidence based on the GWAS data but strong independent evidence in terms of gene expression studies and other prior human or animal genetic work. Conversely, some of the top previous genetic findings in the field,84 such as ADH1C85 (CFG score of 9), GABRA286 (CFG score of 8), as well as AUTS2 (CFG score of 7), CHRM2 and KCNJ6 (CFG scores of 4) have fewer different independent lines of evidence, and thus received a lower CFG prioritization score in our analysis (Supplementary Table S1), although they are clearly involved in alcoholism-related processes. Whereas we cannot exclude that more recently discovered genes have had less hypothesis-driven work performed and thus might score lower on CFG, it is to be noted that the CFG approach integrates predominantly non-hypothesis-driven, discovery-type data sets, such as GWAS data, linkage, quantitative traits loci and, particularly, gene expression. We also cap each line of evidence from an experimental approach (Figure 1), to minimize any ‘popularity’ bias, whereas multiple studies of the same kind are conducted on better-established genes. In the end, it is gene-level reproducibility across multiple approaches and platforms that is built into the approach and gets prioritized most by CFG scoring during the discovery process. Our top results subsequently show good reproducibility and predictive ability in independent cohort testing, the litmus test for any such work.
At the very top of our list of candidate genes for alcoholism, with a CFG score of 13, we have SNCA, a pre-synaptic chaperone that has been reported to be involved in modulating brain plasticity and neurogenesis, as well as neurotransmission, primarily as a brake.87,88 On the pathological side, low levels of SNCA might offer less protection against oxidative stress,89 whereas high levels of SNCA may have a role in neurodegenerative diseases, including in Parkinson disease. SNCA has been identified as a susceptibility gene for alcohol cravings7 and response to alcohol cues.90 The evidence provided by our data and other previous human genetic association studies suggest a genetic rather than purely environmental (alcohol consumption and stress) basis for its alteration in disease, and its potential utility as trait rather than purely state marker.
Alcoholics carry a genetic variant that leads to reduced baseline expression of SNCA.8 SNCA is also downregulated in expression in the frontal cortex and caudate–putamen of inbred alcohol-preferring rats,17 as well as in the brain (amygdala) and blood of our stress-reactive DBP animal model of alcoholism, before exposure to any alcohol. SNCA is upregulated in expression in blood in human alcoholism,12,13 as well as in the blood of monkeys consuming alcohol, and in rats after alcohol administration.3 Thus, it may serve as a blood biomarker. Overall, we may infer that, whereas low levels of SNCA may predispose to cravings for alcohol and consequent alcoholism, possibly mediated through increased neurobiological activity and drive (the SNCA deficit hypothesis), excessive alcohol consumption then increases SNCA expression beyond that seen in non-alcohol-consuming controls, potentially compounding risk for neurodegenerative diseases in individuals that have mutations that lead to its aggregation. This observation is also biologically consistent with the fact that dementia is often observed late in the course of alcohol dependence.
GFAP (glial fibrillary acidic protein), a top candidate gene with a CFG score of 9.5, is an astrocyte intermediate filament-type protein involved in neuron–astrocyte interactions, cell adhesion, process formation and cell–cell communication. It is decreased in expression in post-mortem brain of alcoholics, but increased in expression in brains of animal models of predisposition to alcoholism, before exposure to alcohol (Table 3). This is consistent with a model for increased physiological robustness in individuals predisposed to alcoholism,3 as well as with the neurodegenerative consequences of protracted alcohol use.
DRD2 (dopamine receptor D2), another top candidate gene with a CFG score of 9, has prior human genetic association evidence. It is reduced in expression in the frontal cortex in the human brain from alcoholics, as well as in the DBP animal model before any exposure to alcohol. One possible interpretation would be that lower levels of dopamine receptors are associated with reduced dopaminergic signaling and anhedonia, leading individuals to overcompensate by alcohol and drug abuse. Another interpretation, consistent with the low SNCA and consequently higher neurotransmitter (including dopamine) levels, would be that these individuals are in fact in a compulsive, hyperdopaminergic state, which drives them to hedonic activities and leads to compensatory homeostatic downregulation of their DRD2 receptors. Consistent with this later scenario, mice that have a constitutive knockout of their DRD2 receptors, not because of a hyperdopaminergic state, in fact consume less alcohol,29 unless they are exposed to stress.91
Another top candidate gene, GRM3, is also involved in neurotransmitter signaling. Prior evidence in the field had implicated another metabotropic glutamate receptor, GRM2.92
Other top candidate genes in the panel (MOBP, MBP and MOG) are involved in myelination (Table 3). They are decreased in expression in the prefrontal cortex of human alcoholics, as well as in our stress-reactive DBP animal model of alcoholism, before exposure to any alcohol. Decreased myelination may lead to decreased connectivity. Interestingly, MOBP and MBP are increased in expression in the amygdala in the DBP mice, opposite to the direction of change in the PFC, consistent with a frontal deactivation and a limbic hyperactivity, which could lead to impulsivity.
Epistasis testing of top candidate genes for alcoholism
For the top 11 candidate genes, best P-value SNPs from GWAS1 were used to test for gene–gene interactions in GWAS2 (Supplementary Table S5). Nominally significant interactions were found between SNPs in SNCA and RXRG, DRD2 and SYT1, MOBP and TIMP2. As a caveat, the P-value was not corrected for multiple comparisons. The corresponding genes merit future follow-up work to elucidate the biological and pathophysiological relevance of their interactions.
Pathways and mechanisms
Our pathway analysis (Table 2 and Supplementary Table S2) results are consistent with the accumulating evidence about the role of neuronal excitability and signaling in alcoholism.83,93,94
Overlap with other psychiatric disorders
Despite using lines of evidence for our CFG approach that have to do only with alcoholism, the list of genes identified has a notable overlap at a pathway analysis level (Table 2B and Supplementary Table S2B) and at a gene level (Figures 4 and 5) with other psychiatric disorders. This is a topic of major interest and debate in the field. We demonstrate an overlap between top candidate genes for alcoholism and top candidate genes for schizophrenia, anxiety and bipolar disorder, previously identified by us through CFG (Figure 4), thus providing a possible molecular basis for the frequently observed clinical comorbidity and interdependence between alcoholism and those other major psychiatric disorders, as well as cross-utility of pharmacological agents. Moreover, we tested in alcoholics genetic risk predictive panels for bipolar disorder53 and for schizophrenia54 generated in previous studies by us, and show that they are significantly different in alcoholics versus controls (Figure 6), beyond the overlap in genes with alcohol. There seems to be an increased genetic load for bipolar disorder, consistent with increased drive, and a decreased genetic load for schizophrenia, consistent with increased connectivity before alcohol use. These results led us to develop a heuristic, testable model of alcoholism (Figure 5). Some people may drink to be calm, mitigating the effects of stress and anxiety, some people may drink to be happy, the common drive with bipolar disorder, and some people may drink to be drunk, to disconnect from reality and/or get unstuck from internal obsessions and ruminations.
Genetic risk prediction
Of note, our SNP panels and choice of affected alleles were based solely on analysis of the discovery GWAS, completely independently from the test cohorts. Our results show that a relatively limited and well-defined panel of SNPs identified based on our CFG analysis could differentiate between alcoholism subjects and controls in three independent cohorts. The fact that our genetic testing worked for both alcohol dependence and alcohol abuse suggests that these two diagnostic categories are actually overlapping, supporting the DSM-V reclassification of a single category of alcohol use disorders.
Reproducibility among studies
Our work provides striking evidence for the advantages, reproducibility and consistency of gene-level analyses of data, as opposed to SNP level analyses, pointing to the fundamental issue of genetic heterogeneity at a SNP level. In fact, it may be that the more biologically important a gene is for higher mental functions, the more heterogeneity it has at a SNP level and the more evolutionary divergence, for adaptive reasons. On top of that, CFG provides a way to prioritize genes based on disease relevance, not study-specific effects (that is, fit-to-disease as opposed to fit-to-cohort). Reproducibility of findings across different studies, experimental paradigms and technical platforms is deemed more important (and scored as such by CFG) than the strength of finding in an individual study (for example, P-value in a GWAS).
Potential limitations and confounds
The GWAS study (cohort 1) on which our discovery was based contained males as probands but contained males and females as controls. This was the case for the German test cohort (cohort 2) as well. It is possible that some of the nominally significant SNPs identified in the discovery GWAS have to do with gender differences rather than to alcoholism per se, or at least may have to do with male alcoholism. Stratification across gender and ethnicities may also be a factor in our test cohorts 3 and 4 (Table 1). The issue of possible ethnicity differences in alleles, genes and the consequent neurobiology may need to be explored more in the future, with larger sample sizes, and with environmental and cultural factors taken into account. However, the use of a CFG approach using evidence from other studies of alcoholism, including animal model studies, to prioritize the findings decreases the likelihood that our final top results are ethnicity- or gender-related. Of note, our GRPS predictions separate alcoholics from controls in independent test cohorts, in both genders, and in fact work even better at separating female alcoholics from female controls (Figure 3). Moreover, a series of individual genes from the panel, not just SNCA, separates alcoholics from controls in independent cohorts (Table 5).
The conversion from SNPs to genes as part of our discovery assumed the rule of proximity—that is, an intragenic SNP implicates the gene inside which it falls, or if it falls into an intergenic region, it implicates the most proximal gene to it. That may not be true in reality in all cases, generating potentially false-positives and false-negatives. However, the convergent approach and focus on the top CFG scoring genes reduce the likelihood of false-positives.
The only SNP for SNCA that was present/tested for in cohorts 3 and 4 (rs17015888) was relatively far away upstream (0.13 MB) from SNCA. However, no other known genes are present in that region, SNCA is the closest gene, and the distance is well within the range of known examples of regulatory regions (enhancers). In addition, the risk allele for this SNP (G/G) seems to be the major variant in the population (Supplementary Table S6), suggesting that this allele per se is evolutionarily advantageous, when not coupled with the exogenous ingestion of alcohol.
A relatively large list of genes (n=6085) was implicated by nominally significant SNPs from the discovery GWAS. There is a risk that out of such a large list CFG will find something to prioritize. We have tried to mitigate that by developing an internal score for each gene based on the proportion of SNPs tested in a gene that were nominally significant. Moreover, in the end, we tested the reproducibility and predictive ability of our top findings in multiple independent cohorts, which is the ultimate litmus test for any genetic or biomarker study.
Conclusion
Overall, whereas multiple mechanistic entry points may contribute to alcoholism pathogenesis, it is likely at its core a disease of an exogenous agent (alcohol) modulating different mind domains/dimensions (anxiety, mood and cognition),95 precipitated by environmental stress on a background of genetic vulnerability (Figure 5). The degree to which various mind domains/dimensions are affected in different individuals is a fertile area for future research into subtypes of alcoholism and lends itself to personalization of diagnosis and treatment, by integrating genetic data, blood gene expression biomarker data and clinical data. Lastly, it is important to note that individuals with a predisposition to alcoholism but no exposure to alcohol may in fact have a robust physiology and strong neurobiological drive that can be harnessed for other, more productive endeavors.
References
Schuckit MA . Alcohol-use disorders. Lancet 2009; 373: 492–501.
Niculescu AB, Le-Niculescu H . Convergent Functional Genomics: what we have learned and can learn about genes, pathways, and mechanisms. Neuropsychopharmacology 2010; 35: 355–356.
Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J 2007; 7: 222–256.
Le-Niculescu H, McFarland MJ, Ogden CA, Balaraman Y, Patel S, Tan J et al. Phenomic, convergent functional genomic, and biomarker studies in a stress-reactive genetic animal model of bipolar disorder and co-morbid alcoholism. Am J Med Genet 2008; 147B: 134–166.
Le-Niculescu H, Case NJ, Hulvershorn L, Patel SD, Bowker D, Gupta J et al. Convergent functional genomic studies of omega-3 fatty acids in stress reactivity, bipolar disorder and alcoholism. Transl Psychiatry 2011; 1: e4.
Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 2009; 66: 773–784.
Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J et al. Association of alcohol craving with alpha-synuclein (SNCA). Alcohol Clin Exp Res 2007; 31: 537–545.
Janeczek P, Mackay RK, Lea RA, Dodd PR, Lewohl JM . Reduced expression ofalpha-synuclein in alcoholic brain: influence of SNCA-Rep1 genotype. Addict Biol 2014; 19: 509–515.
Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA . Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem 2002; 81: 802–813.
Lewohl JM, Van Dyk DD, Craft GE, Innes DJ, Mayfield RD, Cobon G et al. The application of proteomics to the human alcoholic brain. Ann N Y Acad Sci 2004; 1025: 14–26.
Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA . Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res 2000; 24: 1873–1882.
Taraskina AE, Filimonov VA, Kozlovskaya YA, Morozova MN, Gaschin DV, Schwarzman AL . High level of alpha-synuclein mRNA in peripheral lymphocytes of patients with alcohol dependence syndrome. Bull exp Biol Med 2008; 146: 609–611.
Bonsch D, Reulbach U, Bayerlein K, Hillemacher T, Kornhuber J, Bleich S . Elevated alpha synuclein mRNA levels are associated with craving in patients with alcoholism. Biol Psychiatry 2004; 56: 984–986.
Bonsch D, Greifenberg V, Bayerlein K, Biermann T, Reulbach U, Hillemacher T et al. Alpha-synuclein protein levels are increased in alcoholic patients and are linked to craving. Alcohol Clin Exp Res 2005; 29: 763–765.
Bonsch D, Lenz B, Kornhuber J, Bleich S . DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport 2005; 16: 167–170.
Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA et al. alpha-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc Natl Acad Sci USA 2003; 100: 4690–4695.
Liang T, Kimpel MW, McClintick JN, Skillman AR, McCall K, Edenberg HJ et al. Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats. Genome Biol 2010; 11: R11.
Walker SJ, Grant KA . Peripheral blood alpha-synuclein mRNA levels are elevated in cynomolgus monkeys that chronically self-administer ethanol. Alcohol 2006; 38: 1–4.
Dick DM, Aliev F, Bierut L, Goate A, Rice J, Hinrichs A et al. Linkage analyses of IQ in the collaborative study on the genetics of alcoholism (COGA) sample. Behav Genet 2006; 36: 77–86.
Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD . Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem 2004; 90: 1050–1058.
Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ et al. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol 2007; 41: 95–132.
Ponce G, Perez-Gonzalez R, Aragues M, Palomo T, Rodriguez-Jimenez R, Jimenez-Arriero MA et al. The ANKK1 kinase gene and psychiatric disorders. Neurotoxic Res 2009; 16: 50–59.
Dick DM, Wang JC, Plunkett J, Aliev F, Hinrichs A, Bertelsen S et al. Family-based association analyses of alcohol dependence phenotypes across DRD2 and neighboring gene ANKK1. Alcohol Clin Exp Res 2007; 31: 1645–1653.
Kraschewski A, Reese J, Anghelescu I, Winterer G, Schmidt LG, Gallinat J et al. Association of the dopamine D2 receptor gene with alcohol dependence: haplotypes and subgroups of alcoholics as key factors for understanding receptor function. Pharmacogenet Genomics 2009; 19: 513–527.
Munafo MR, Matheson IJ, Flint J . Association of the DRD2 gene Taq1A polymorphism and alcoholism: a meta-analysis of case-control studies and evidence of publication bias. Mol Psychiatry 2007; 12: 454–461.
Joe KH, Kim DJ, Park BL, Yoon S, Lee HK, Kim TS et al. Genetic association of DRD2 polymorphisms with anxiety scores among alcohol-dependent patients. Biochem Biophys Res Commun 2008; 371: 591–595.
Noble EP, Blum K, Ritchie T, Montgomery A, Sheridan PJ . Allelic association of the D2 dopamine receptor gene with receptor-binding characteristics in alcoholism. Arch Gen Psychiatry 1991; 48: 648–654.
Volkow ND, Wang GJ, Fowler JS, Logan J, Hitzemann R, Ding YS et al. Decreases in dopamine receptors but not in dopamine transporters in alcoholics. Alcohol Clin Exp Res 1996; 20: 1594–1598.
Phillips TJ, Brown KJ, Burkhart-Kasch S, Wenger CD, Kelly MA, Rubinstein M et al. Alcohol preference and sensitivity are markedly reduced in mice lacking dopamine D2 receptors. Nat Neurosci 1998; 1: 610–615.
Zhu X, Cooper R, Kan D, Cao G, Wu X . A genome-wide linkage and association study using COGA data. BMC Genet 2005; 6 (Suppl 1): S128.
Wang S, Huang S, Liu N, Chen L, Oh C, Zhao H . Whole-genome linkage analysis in mapping alcoholism genes using single-nucleotide polymorphisms and microsatellites. BMC Genet 2005; 6 (Suppl 1): S28.
McClintick JN, Xuei X, Tischfield JA, Goate A, Foroud T, Wetherill L et al. Stress-response pathways are altered in the hippocampus of chronic alcoholics. Alcohol 2013; 47: 505–515.
Sommer W, Arlinde C, Heilig M . The search for candidate genes of alcoholism: evidence from expression profiling studies. Addict Biol 2005; 10: 71–79.
Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K et al. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res 2005; 80: 529–538.
Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR et al. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J 2013; 13: 286–296.
Zoeller RT, Butnariu OV, Fletcher DL, Riley EP . Limited postnatal ethanol exposure permanently alters the expression of mRNAS encoding myelin basic protein and myelin-associated glycoprotein in cerebellum. Alcohol Clin Exp Res 1994; 18: 909–916.
Chiappelli F, Taylor AN, Espinosa de los Monteros A, de Vellis J . Fetal alcohol delays the developmental expression of myelin basic protein and transferrin in rat primary oligodendrocyte cultures. Int J Dev Neurosci 1991; 9: 67–75.
Bichenkov E, Ellingson JS . Ethanol alters the expressions of c-Fos and myelin basic protein in differentiating oligodendrocytes. Alcohol 2009; 43: 627–634.
Weng J, Symons MN, Singh SM . Ethanol-responsive genes (Crtam, Zbtb16, and Mobp) located in the alcohol-QTL region of chromosome 9 are associated with alcohol preference in mice. Alcohol Clin Exp Res 2009; 33: 1409–1416.
Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T et al. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav 2009; 94: 131–147.
Treadwell JA, Singh SM . Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res 2004; 29: 357–369.
Bergen AW, Yang XR, Bai Y, Beerman MB, Goldstein AM, Goldin LR . Genomic regions linked to alcohol consumption in the Framingham Heart Study. BMC genet 2003; 4 (Suppl 1): S101.
Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res 2000; 24: 933–945.
McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hauser SR, Edenberg HJ et al. Changes in gene expression within the ventral tegmental area following repeated excessive binge-like alcohol drinking by alcohol-preferring (P) rats. Alcohol 2013; 47: 367–380.
Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W . A genome wide search for alcoholism susceptibility genes. Am J Med Genet 2004; 128: 102–113.
Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem 2005; 93: 359–370.
Kabbaj M, Evans S, Watson SJ, Akil H . The search for the neurobiological basis of vulnerability to drug abuse: using microarrays to investigate the role of stress and individual differences. Neuropharmacology 2004; 47 (Suppl 1): 111–122.
Pignataro L, Miller AN, Ma L, Midha S, Protiva P, Herrera DG et al. Alcohol regulates gene expression in neurons via activation of heat shock factor 1. J Neurosci 2007; 27: 12957–12966.
Varodayan FP, Pignataro L, Harrison NL . Alcohol induces synaptotagmin 1 expression in neurons via activation of heat shock factor 1. Neuroscience 2011; 193: 63–71.
Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology 2006; 31: 1574–1582.
Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P et al. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol 2012; 17: 171–180.
Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 2014; 19: 41–49.
Patel SD, Le-Niculescu H, Koller DL, Green SD, Lahiri DK, McMahon FJ et al. Coming to grips with complex disorders: genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am J Med Genet 2010; 153B: 850–877.
Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry 2012; 17: 887–905.
Niculescu AB, Le-Niculescu H . The P-value illusion: how to improve (psychiatric) genetic studies. Am J Med Genet 2010; 153B: 847–849.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Med Genet 2007; 81: 559–575.
Yang J, Lee SH, Goddard ME, Visscher PM . GCTA: a tool for genome-wide complex trait analysis. Am J Med Genet 2011; 88: 76–82.
Edwards S, Baynes BB, Carmichael CY, Zamora-Martinez ER, Barrus M, Koob GF et al. Traumatic stress reactivity promotes excessive alcohol drinking and alters the balance of prefrontal cortex-amygdala activity. Transl Psychiatry 2013; 3: e296.
Koob GF, Buck CL, Cohen A, Edwards S, Park PE, Schlosburg JE et al. Addiction as a stress surfeit disorder. Neuropharmacology 2014; 76 Pt B: 370–382.
Schank JR, Ryabinin AE, Giardino WJ, Ciccocioppo R, Heilig M . Stress-related neuropeptides and addictive behaviors: beyond the usual suspects. Neuron 2012; 76: 192–208.
Niculescu AB 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR . Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics 2000; 4: 83–91.
Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet 2009; 150B: 155–181.
Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR et al. Genome-wideassociation study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry 2014; 19: 41–49.
DeMar JC Jr, Ma K, Bell JM, Igarashi M, Greenstein D, Rapoport SI . One generation of n-3 polyunsaturated fatty acid deprivation increases depression and aggression test scores in rats. J Lipid Res 2006; 47: 172–180.
Rao JS, Ertley RN, Lee HJ, DeMar JC Jr, Arnold JT, Rapoport SI et al. n-3 polyunsaturated fatty acid deprivation in rats decreases frontal cortex BDNF via a p38 MAPK-dependent mechanism. Mol Psychiatry 2007; 12: 36–46.
Zanarini MC, Frankenburg FR . omega-3 Fatty acid treatment of women with borderline personality disorder: a double-blind, placebo-controlled pilot study. Am J Psychiatry 2003; 160: 167–169.
Lin PY, Huang SY, Su KP . A meta-analytic review of polyunsaturated fatty acid compositions in patients with depression. Biol Psychiatry 2010; 68: 140–147.
Collins MA, Moon KH, Tajuddin N, Neafsey EJ, Kim HY . Docosahexaenoic acid (DHA) prevents binge ethanol-dependent aquaporin-4 elevations while inhibiting neurodegeneration: experiments in rat adult-age entorhino-hippocampal slice cultures. Neurotox Res 2013; 23: 105–110.
Patten AR, Brocardo PS, Christie BR . Omega-3 supplementation can restore glutathione levels and prevent oxidative damage caused by prenatal ethanol exposure. J Nutr Biochem 2012; 24: 760–769.
McNamara RK . DHA deficiency and prefrontal cortex neuropathology in recurrent affective disorders. J Nutr 2010; 140: 864–868.
McNamara RK, Jandacek R, Rider T, Tso P, Stanford KE, Hahn CG et al. Deficits in docosahexaenoic acid and associated elevations in the metabolism of arachidonic acid and saturated fatty acids in the postmortem orbitofrontal cortex of patients with bipolar disorder. Psychiatry Res 2008; 160: 285–299.
Sublette ME, Hibbeln JR, Galfalvy H, Oquendo MA, Mann JJ . Omega-3 polyunsaturated essential fatty acid status as a predictor of future suicide risk. Am J Psychiatry 2006; 163: 1100–1102.
Lewis MD, Hibbeln JR, Johnson JE, Lin YH, Hyun DY, Loewke JD . Suicide deaths of active-duty US military and omega-3 fatty-acid status: a case-control comparison. J Clin Psychiatry 2011; 72: 1585–1590.
Stoll AL, Severus WE, Freeman MP, Rueter S, Zboyan HA, Diamond E et al. Omega 3 fatty acids in bipolar disorder: a preliminary double-blind, placebo-controlled trial. Arch Gen Psychiatry 1999; 56: 407–412.
Parker G, Gibson NA, Brotchie H, Heruc G, Rees AM, Hadzi-Pavlovic D . Omega-3 Fatty acids and mood disorders. Am J Psychiatry 2006; 163: 969–978.
Osher Y, Belmaker RH . Omega-3 fatty acids in depression: a review of three studies. CNS Neurosci Ther 2009; 15: 128–133.
Clayton EH, Hanstock TL, Hirneth SJ, Kable CJ, Garg ML, Hazell PL . Reduced mania and depression in juvenile bipolar disorder associated with long-chain omega-3 polyunsaturated fatty acid supplementation. Eur J Clin Nutr 2009; 63: 1037–1040.
Peet M, Stokes C . Omega-3 fatty acids in the treatment of psychiatric disorders. Drugs 2005; 65: 1051–1059.
Berger GE, Wood SJ, Wellard RM, Proffitt TM, McConchie M, Amminger GP et al. Ethyl-eicosapentaenoic acid in first-episode psychosis. A 1H-MRS study. Neuropsychopharmacology 2008; 33: 2467–2473.
Amminger GP, Schafer MR, Papageorgiou K, Klier CM, Cotton SM, Harrigan SM et al. Long-chain omega-3 fatty acids for indicated prevention of psychotic disorders: a randomized, placebo-controlled trial. Arch Gen Psychiatry 2010; 67: 146–154.
Kurian SM, Le-Niculescu H, Patel SD, Bertram D, Davis J, Dike C et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol Psychiatry 2009; 16: 37–58.
Yan J, Aliev F, Webb BT, Kendler KS, Williamson VS, Edenberg HJ et al. Using geneticinformation from candidate gene and genome-wide association studies in risk prediction for alcohol dependence. Addict Biol advance online publication, 30 January 2013; doi:10.1111/adb.12035 (e-pub ahead of print).
Kos MZ, Yan J, Dick DM, Agrawal A, Bucholz KK, Rice JP et al. Common biological networks underlie genetic risk for alcoholism in African- and European-American populations. Genes Brain Behavior 2013; 12: 532–542.
Edenberg HJ, Foroud T . Genetics and alcoholism. Nat Rev Gastroenterol Hepatol 2013; 10: 487–494.
Biernacka JM, Geske JR, Schneekloth TD, Frye MA, Cunningham JM, Choi DS et al. Replication of genome wide association studies of alcohol dependence: support for association with variation in ADH1C. PLoS ONE 2013; 8: e58798.
Villafuerte S, Strumba V, Stoltenberg SF, Zucker RA, Burmeister M . Impulsiveness mediates the association between GABRA2 SNPs and lifetime alcohol problems. Genes Brain Behavior 2013; 12: 525–531.
Winner B, Regensburger M, Schreglmann S, Boyer L, Prots I, Rockenstein E et al. Role of alpha-synuclein in adult neurogenesis and neuronal maturation in the dentate gyrus. J Neurosci 2012; 32: 16906–16916.
Lundblad M, Decressac M, Mattsson B, Bjorklund A . Impaired neurotransmission caused by overexpression of alpha-synuclein in nigral dopamine neurons. Proc Natl Acad Sci USA 2012; 109: 3213–3219.
Musgrove RE, King AE, Dickson TC . alpha-Synuclein protects neurons from apoptosis downstream of free-radical production through modulation of the MAPK signalling pathway. Neurotox Res 2013; 23: 358–369.
Wilcox CE, Claus ED, Blaine SK, Morgan M, Hutchison KE . Genetic variation in the alpha synuclein gene (SNCA) is associated with BOLD response to alcohol cues. J Stud Alcohol Drugs 2013; 74: 233–244.
Delis F, Thanos PK, Rombola C, Rosko L, Grandy D, Wang GJ et al. Chronic mild stress increases alcohol intake in mice with low dopamine D2 receptor levels. Behav Neurosci 2013; 127: 95–105.
Zhou Z, Karlsson C, Liang T, Xiong W, Kimura M, Tapocik JD et al. Loss of metabotropic glutamate receptor 2 escalates alcohol consumption. Proc Natl Acad Sci USA 2013; 110: 16963–16968.
Heilig M, Goldman D, Berrettini W, O'Brien CP . Pharmacogenetic approaches to the treatment of alcohol addiction. Nat Rev Neurosci 2011; 12: 670–684.
Zhao Z, Guo AY, van den Oord EJ, Aliev F, Jia P, Edenberg HJ et al. Multi-species data integration and gene ranking enrich significant results in an alcoholism genome-wide association study. BMC Genomics 2012; 13 (Suppl 8): S16.
Niculescu AB 3rd, Schork NJ, Salomon DR . Mindscape: a convergent perspective on life, mind, consciousness and happiness. J Affect Disord 2010; 123: 1–8.
Acknowledgements
This work is, in essence, a field-wide collaboration. We would like to acknowledge our debt of gratitude for the efforts and results of the many other groups, cited in our paper, who have conducted and published studies (genetic and gene expression) in alcoholism. With their arduous and careful work, a convergent approach, such as ours, is possible. We would particularly like to thank the subjects who volunteered to participate in these studies. Without their generous contribution, such work to advance the understanding of alcoholism and help others would not be possible. This work was supported by an NIH Directors’ New Innovator Award (1DP2OD007363) and a VA Merit Award (1I01CX000139-01) to ABN, as well as by NIH grants R01 DA12690, R01 DA12849, R01 AA11330, R01 and AA017535 to JG and collaborators, and by grant FKZ 01GS08152 from the National Genome Research Network of the German Federal Ministry of Education and Research to MR and collaborators.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest directly related to this work. Although not directly relevant to this work, HRK has been a consultant or advisory board member with Alkermes, Lilly, Lundbeck, Pfizer, and Roche. He has also received honoraria from the Alcohol Clinical Trials Initiative (ACTIVE) of the American Society of Clinical Psychopharmacology, which is supported by Lilly, Lundbeck, AbbVie and Pfizer.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Levey, D., Le-Niculescu, H., Frank, J. et al. Genetic risk prediction and neurobiological understanding of alcoholism. Transl Psychiatry 4, e391 (2014). https://doi.org/10.1038/tp.2014.29
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2014.29
This article is cited by
-
Subjective responses to amphetamine in young adults with previous mood elevation experiences
Psychopharmacology (2019)
-
Mood, stress and longevity: convergence on ANK3
Molecular Psychiatry (2016)
-
Molecular signatures associated with cognitive deficits in schizophrenia: a study of biopsied olfactory neural epithelium
Translational Psychiatry (2016)