Abstract
To follow-up loci discovered by the International Genomics of Alzheimer’s Disease Project, we attempted independent replication of 19 single nucleotide polymorphisms (SNPs) in a large Spanish sample (Fundació ACE data set; 1808 patients and 2564 controls). Our results corroborate association with four SNPs located in the genes INPP5D, MEF2C, ZCWPW1 and FERMT2, respectively. Of these, ZCWPW1 was the only SNP to withstand correction for multiple testing (P=0.000655). Furthermore, we identify TRIP4 (rs74615166) as a novel genome-wide significant locus for Alzheimer’s disease risk (odds ratio=1.31; confidence interval 95% (1.19–1.44); P=9.74 × 10−9).
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a complex multifactorial neuropsychiatric disorder whose etiology involves both environmental and genetic factors. The major genetic risk factor for AD is the apolipoprotein E4 (APOE4) allele.1 For 17 years after its discovery, this remained the only confirmed genetic risk factor for the disorder. However, subsequent meta-analyses of genome-wide association studies identified further genetic risk loci. These include signals close to, or within, candidate genes such as CLU,2,3 PICALM,2 CR1,3 BIN1,4 ABCA7,5 EPHA1,5,6 CD33,5,6 CD2AP5,6 and ATP5H/KCTD2,7 as well as the MS4A gene cluster.5,6,8
The International Genomics of Alzheimer’s Disease Project (IGAP) is the largest genetic epidemiology investigation of AD risk to date. In 2013, the IGAP reported a mega meta-analysis, which was divided into a discovery step (stage 1) and a replication step (stage 2). This mega meta-analysis comprised 74 046 samples, including those of 25 580 AD cases, and identified 11 new loci, thus doubling the number of genome-wide significant loci reported for AD.9 The analysis also identified 13 suggestive loci. These findings may serve as the starting point for novel discoveries in future AD genomics studies.
Four of the 11 genome-wide significant loci in the IGAP analyses reached significance in stage 1 (rs8093731 DSG2; rs28834970 PTK2B; rs11218343 SORL1; rs10498633 SLC24A4). The remaining seven only reached genome-wide significance in stage 2—that is, after the inclusion of the replication sample (rs35349669 INPP5D; rs190982 MEF2C; rs2718058 NME8; rs1476679 ZCWPW1; rs10838725 CELF1; rs17125944 FERMT2, and rs7274581 CASS4). Replication of the IGAP findings in an independent series is therefore warranted. The present report describes the follow-up of 10 novel and nine suggestive IGAP loci using subjects drawn from the Spanish Fundació ACE cohort.7
Materials and methods
Patients and controls
The present study involved 4372 individuals. These included 1 808 patients with a possible or probable diagnosis of AD, as assigned by a neurologist,8,10 and 2564 unrelated healthy controls from the Spanish general population who were selected from the Neocodex bio-bank.11 The AD cases were recruited consecutively from three centers: Barcelona (n=1627); Madrid (n=161); and Murcia (n=20). None of these AD patients had been included in the IGAP replication analyses. To avoid the issue of population stratification, all cases and controls were of Spanish ancestry, which was defined as a history of two generations of registered Spanish ancestors. The demographic characteristics of the Fundació ACE participants are described elsewhere.7 Written informed consent was obtained from all participants, or from their legal representatives when necessary. The study was approved by the respective ethics committees, and was performed in accordance with the Code of Ethical Principles for Medical Research Involving Human Subjects of the World Medical Association.
DNA isolation and genotyping
DNA was extracted using ‘Magnapure’ technology (Roche Diagnostics, Mannheim, Germany). Twenty-five single nucleotide length polymorphisms (SNPs) with an AD-association risk of below P<10−6 in the IGAP consortium study were selected for replication. The primer molecules for the multiplex reaction were designed using the Assay Design Suite tool (www.mysequenom.com, Sequenom, San Diego, CA, USA). Assay designs were successful for 21 of the 25 selected variants. Four SNPs (rs72807343 (SQSTM1); rs9271192 (HLA-DRB5/HLA-DRB1); rs2337406 (IGH@); and chr17:61,538,148 (ACE)) were rejected during this phase due to technical problems. Primer sequences and assay conditions for the genotyped SNPs are available upon request.
Quality control
A total of 1808 AD patients and 2564 controls were genotyped for 21 SNPs using Sequenom's Mass Array System (Sequenom) and iPlex Gold reagents in accordance with the manufacturer's instructions. Only SNPs with a call rate of ⩾95% and a Hardy–Weinberg equilibrium P-value of >0.01 in the whole data set were included in the subsequent analyses (Supplementary Table 1). All SNP major and minor alleles and allelic frequencies obtained in the Fundació ACE data set were fully consistent with those reported by the IGAP consortium (Table 1). The overall conversion rate was 96.7%. The SNPs rs10751667 (AP2A2) and rs10838725 (CELF1) failed quality control and were excluded from the statistical analyses. The 19 successfully genotyped SNPs and their status in the IGAP analyses (that is, genome-wide association studies significant or suggestive) are specified in Table 1.
Statistical analysis
Genetic association analyses and calculation of allelic frequencies and Hardy–Weinberg equilibrium were conducted using the online tool at the TUM Helmholtz Center (Munich, Germany; http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). Briefly, tests adapted from those of Sasieni12 were used. Age- and sex-adjusted binary logistic regression analyses were performed using SPSS 15.0 software (SPSS, Chicago, IL, USA). In addition, Mantel–Haenzsel-stratified analyses were conducted according to gender and the presence or absence of the APOE-ɛ4 allele (Supplementary Tables 2 and 3, respectively). Breslow-day tests were conducted to measure the significance of SNP × APOE and SNP × gender interactions. All SNP calculations were double-checked using PLINK or INTERSNP software.13,14 Meta-analyses were conducted using the PLINK software. All results were doubled-checked using Ken Rothman’s Episheet spreadsheet and PLINK (for details see http://pngu.mgh.harvard.edu/~purcell/plink/; http://krothman.hostbyet2.com/episheet.xls). Power calculations were performed using Episheet.
Results and Discussion
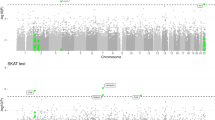
In the present replication effort, a nominally significant signal (P<0.05) was detected for six of the 19 investigated SNPs: rs35349669 at INPP5D (P=0.023); rs190982 at MEF2C (P=0.0062); rs1476679 at ZCWPW1 (P=0.00065); rs17125944 at FERMT2 (P=0.018); rs8035452 at SPPL2A (P=0.031); and rs74615166 at TRIP4 (P=0.0032) (Table 1). Of these, rs1476679 at the ZCWPW1 locus, which had shown genome-wide significance in stage 2 of the IGAP analyses, was the only SNP to withstand correction for multiple testing (P=0.000655). However, the observed inflation factor for χ2 was λ=4.7. As in genome-wide analyses, the λ-value was computed as the median of the χ2-test statistics obtained for the 19 investigated SNPs. Under the null hypothesis of no association, the expected λ value is 1 in the absence of true association. In our study, however, the λ value is 4.7, which is a strong sign of an overall increased degree of significant associations for the SNPs investigated in the Spanish cohort. In the present context, the λ value is considered to indicate the overall degree of positive associations for a small set of SNPs, in contrast to the genome-wide setting, where it is used as an indicator of residual inflation caused by spurious association. Thus, the present observations in our study are unlikely to represent chance findings. Furthermore, four of these five nominally significant association signals displayed the same effect direction as that reported by the IGAP (Table 1). The exception was the marker rs8035452. The IGAP reported this as a suggestive signal. However, an effect in the opposite direction was found in the Fundació ACE data set. This observation might reflect a lack of power in our data set to detect this signal. Alternatively, the original finding may represent a false-positive.
No significant association was found for three of the four genome-wide significant loci detected during IGAP Stage 1 (rs11218343 at SORL1; rs10498633 at SLC24A4; rs8093731 at DSG2). However, since the effect sizes and directions of these three loci were fully consistent with those reported by the IGAP, our failure to replicate them may have been attributable to a lack of power.
In total, seven of the 10 investigated genome-wide significant loci from the IGAP displayed a consistent effect in the present data set. The non-consistent effects observed for the NME8, PTKB2, and CASS4 signals may have been attributable to a lack of power. It should be noted that, although our series may appear underpowered compared with the IGAP data set, the present study had on average a power of 45% to detect each of the genome-wide association studies significant signals reported by the IGAP.
The results of the APOE and gender-adjusted stratified analyses suggested that for most of the 19 investigated SNPs, APOE status and gender had little impact on effect size or the association results (Supplementary Table 3). Interestingly, nominal P-values for APOE interaction were obtained for two nonsignificant SNPs in our series (rs7295246 ADAMST20; and rs7225151 SCIMP). Both loci were reported as being suggestive by the IGAP. The results of the present stratification analyses support the hypothesis that these two loci represent susceptibility factors in only a fraction of AD patients, and that their effects are dependent upon the APOE-ɛ4 genotype. This observation may facilitate determination of their role in AD development in future studies (Supplementary Table 3).
Of the nine suggestive loci proposed by the IGAP, only one SNP was significant in the present analyses (Table 1). The statistically significant signal was obtained for rs74615166 at the thyroid receptor interacting protein gene 4, TRIP4, locus (odd ratio=1.519 (1.148–2.012), P=0.0032). This variant had a minor allele frequency of 0.02 in both the IGAP and the Fundació ACE (Table 1). Interestingly, a larger effect size was observed in the Fundació ACE data set than in the IGAP. However, an advantage of the present analyses was that this SNP was genotyped directly, whereas the IGAP had to rely in part on imputed genotypes. Since imputation for rarer variants is more difficult, this might explain why a stronger effect was observed in the present cohort. The present findings for the nine suggestive IGAP signals may indicate that these loci have a weaker effect on AD risk than the genome-wide significant SNPs. As a direct consequence, the power to detect them using our data set is relatively low (33% on average for suggestive signals). However, our data set had a >99.9% power to detect at least one suggestive locus (0.33),9 thus explaining the results for the TRIP4 locus.
Meta-analysis of the present results with the IGAP meta-analysis data identified TRIP4 as a novel genome-wide significant locus. The new susceptibility AD SNP is located within the eleventh intron of TRIP4 (15q22.31; rs74615166; OR=1.31 (1.17–1.42), P=9.74 × 10−9; Table 1). According to publicly available databases (genome.cse.ucsc.edu), TRIP4 is highly expressed in the immune system and has been detected in various tissues, including the brain. Research suggests that TRIP4 is a component of the nuclear receptor-coupled co-activation machinery that enables or disables DNA transcription.15 A homolog of the TRIP4 gene in Caenorhabditis elegans showed elevated transcript levels in aged or starved adults, which suggests that TRIP4 has a role in cellular maintenance or survival.16 The TRIP class of proteins show thyroid hormone-dependent interaction with their receptors, and the association between TRIP4 and AD risk may partly explain previous findings of an association between low thyroid-stimulating hormone levels in clinically euthyroid subjects and increased AD risk.17 TRIP proteins show a similar ligand-dependent interaction with the retinoid X receptor. This is of interest, since a recent AD mouse model study reported that administration of the retinoid X receptor agonist bexarotene resulted, within hours, in enhanced clearance of soluble Aβ.18 Besides TRIP4, the linkage disequilibrium block that contains rs74615166 includes several other candidate genes, such as CSNK1G1. CSNK1G1 is a member of the CK-1 family, and its gene product has been implicated in the amyloid cascade.19
A major limitation of the present study was the lack of power to confirm all true associations. Therefore, our negative results cannot be interpreted as confirmation of a lack of association for the respective SNPs, which remain putative susceptibility loci for AD. A fraction of the suggestive SNPs reported by the IGAP may be genuine, and these SNPs warrant further investigation.
The results obtained for TRIP4 underscore the importance of follow-up and comprehensive replication of consortia results. Further genotyping and re-sequencing efforts to investigate TRIP4 and the other IGAP loci are underway in order to elucidate the role of TRIP4 in AD risk and corroborate further genuine signals. Further studies are now warranted to identify the functional mechanism underlying the association between TRIP4 and AD.
References
Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry 2011; 16: 903–907.
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 2009; 41: 1088–1093.
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M et al Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009; 41: 1094–1099.
Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 2010; 303: 1832–1840.
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011; 43: 429–435.
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011; 43: 436–441.
Boada M, Antunez C, Ramirez-Lorca R, Destefano AL, Gonzalez-Perez A, Gayan J et al ATP5H/KCTD2 locus is associated with Alzheimer's disease risk. Mol Psychiatry advance online publication, 16 July 2013; doi: 10.1038/mp.2013.86; e-pub ahead of print.
Antunez C, Boada M, Gonzalez-Perez A, Gayan J, Ramirez-Lorca R, Marin J et al. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer's disease. Genome Med 2011; 3: 33.
Lambert JC, Ibrahim-Verbaas CA, Denise Harold D, Adam C Naj AC, Sims R, Bellenguez C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–8.
Corey-Bloom J, Galasko D, Hofstetter CR, Jackson JE, Thal LJ . Clinical features distinguishing large cohorts with possible AD, probable AD, and mixed dementia. J Am Geriatr Soc 1993; 41: 31–37.
Gayan J, Galan JJ, Gonzalez-Perez A, Saez ME, Martinez-Larrad MT, Zabena C et al. Genetic structure of the Spanish population. BMC Genomics 2010; 11: 326.
Sasieni PD . From genotypes to genes: doubling the sample size. Biometrics 1997; 53: 1253–1261.
Herold C, Steffens M, Brockschmidt FF, Baur MP, Becker T . INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics 2009; 25: 3275–3281.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Jung DJ, Sung HS, Goo YW, Lee HM, Park OK, Jung SY et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol Cell Biol 2002; 22: 5203–5211.
Cherkasova V, Ayyadevara S, Egilmez N, Shmookler Reis R . Diverse Caenorhabditis elegans genes that are upregulated in dauer larvae also show elevated transcript levels in long-lived, aged, or starved adults. J Mol Biol 2000; 300: 433–448.
Tan ZS, Beiser A, Vasan RS, Au R, Auerbach S, Kiel DP et al. Thyroid function and the risk of Alzheimer disease: the Framingham Study. Arch Intern Med 2008; 168: 1514–1520.
Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE et al. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science 2012; 335: 1503–1506.
Flajolet M, He G, Heiman M, Lin A, Nairn AC, Greengard P . Regulation of Alzheimer's disease amyloid-beta formation by casein kinase I. Proc Natl Acad Sci USA 2007; 104: 4159–4164.
Acknowledgements
We thank all patients and controls for their participation in this project. We thank all of the investigators from the International Genomics of Alzheimer Project (IGAP) consortium for their close collaboration and intellectual input, and for sharing their pre-publication data in order to permit rapid completion of this research work. The IGAP comprises four major Alzheimer’s disease research groups: the Alzheimer Disease Genetics Consortium (ADGC); the Neurology Working Group of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE); the European Initiative for Alzheimer Disease (EADI); and the Genetic and Environmental Risk in Alzheimer's Disease (GERAD). We are indebted to Trinitat Port-Carbó and her family for their support of the Fundació ACE research programs. Fundació ACE collaborates with the Centro de Investigación Biomédica en Red sobre Enfermedades Neurodegenerativas (CIBERNED, Spain), and is one of the participating centers of the Dementia Genetics Spanish Consortium (DEGESCO).The Diabetes Research Laboratory, Biomedical Research Foundation at University Hospital Clínico San Carlos received support from CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM). CIBERDEM and CIBERNED are Instituto de Salud Carlos III ISCIII Projects. Agustín Ruiz is supported by grant PI13/02434 (Acción Estratégica en Salud. Instituto de Salud Carlos III (ISCIII). Ministerio de Economía y Competitividad, Spain). Tim Becker and Markus M Nöthen are members of the DFG-funded Excellence Cluster ImmunoSensation. This study was funded in part by the National institute of Health (NIH) project AG033193 to CHARGE. This publication was funded in part by the German Federal Ministry of Education and Research (grants KND: 01GI0102, 01GI0420, 01GI0422, 01GI0423, 01GI0429, 01GI0431, 01GI0433, 01GI0434; grants KNDD: 01GI0710, 01GI0711, 01GI0712, 01GI0713, 01GI0714, 01GI0715, 01GI0716, 01ET1006B).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
A complete list of members of IGAP and their affiliations appears in the Supplementary note.
Supplementary Information accompanies the paper on the Translational Psychiatry website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Ruiz, A., Heilmann, S., Becker, T. et al. Follow-up of loci from the International Genomics of Alzheimer’s Disease Project identifies TRIP4 as a novel susceptibility gene. Transl Psychiatry 4, e358 (2014). https://doi.org/10.1038/tp.2014.2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2014.2
Keywords
This article is cited by
-
The neuroimmune axis of Alzheimer’s disease
Genome Medicine (2023)
-
Unraveling the phenotypic and genomic background of behavioral plasticity and temperament in North American Angus cattle
Genetics Selection Evolution (2023)
-
Progress on the roles of MEF2C in neuropsychiatric diseases
Molecular Brain (2022)
-
Identification of a novel mutation in MEF2C gene in an atypical patient with frontotemporal lobar degeneration
Neurological Sciences (2022)
-
Genomics and Functional Genomics of Alzheimer's Disease
Neurotherapeutics (2022)