Abstract
Currently used antidepressants elevate monoamine levels in the synaptic cleft. There is good reason to assume that this is not the only source for antidepressant therapeutic activities and that secondary downstream effects may be relevant for alleviating symptoms of depression. We attempted to elucidate affected biochemical pathways downstream of monoamine reuptake inhibition by interrogating metabolomic profiles in DBA/2Ola mice after chronic paroxetine treatment. Metabolomic changes were investigated using gas chromatography-mass spectrometry profiling and group differences were analyzed by univariate and multivariate statistics. Pathways affected by antidepressant treatment were related to energy metabolism, amino acid metabolism and hormone signaling. The identified pathways reveal further antidepressant therapeutic action and represent targets for drug development efforts. A comparison of the central nervous system with blood plasma metabolite alterations identified GABA, galactose-6-phosphate and leucine as biomarker candidates for assessment of antidepressant treatment effects in the periphery.
Similar content being viewed by others
Introduction
Major depression is a severe and life-threatening disease with a population prevalence of 17%1 and is projected to be the second leading contributor to the global burden of disease (DALYs) by the year 2020.2 At present, selective serotonin reuptake inhibitors, selective noradrenaline reuptake inhibitors and monoamine oxidase inhibitors are the most commonly prescribed antidepressants.3, 4 Shortcomings of current antidepressant treatments include a delayed onset of therapeutic action, adverse side effects and response in only a subset of patients.4, 5
The elevation of serotonin or noradrenaline levels by tricyclic and currently used antidepressants had led to the monoamine deficiency hypothesis of depression6, 7 with clinical research over the past 40 years suggesting that increased monoaminergic neurotransmission is essential for antidepressant efficiency.6 However, contrary to this hypothesis, monoamine depletion does not induce depression in healthy humans.8 Moreover, a study by Nickel et al.9 found that paroxetine, a serotonin reuptake inhibitor and tianeptine, a serotonin reuptake enhancer are both effective antidepressants. A common downstream effect of both drugs was to normalize hypothalamic-pituitary-adrenal (HPA) axis reactivity. These findings indicate that opposite effects on the serotonin system may lead to common downstream pathway alterations. The most parsimonious explanation is that monoamine elevation per se is not the only source for antidepressant activity but secondary downstream effects may also be involved in the alleviation of depressive symptoms.10, 11, 12 Therefore, it is tempting to speculate that for improved antidepressant treatment it would be beneficial to pharmacologically interfere with these downstream mechanisms directly. Proposed antidepressant downstream effects include neurogenesis,13, 14 strengthened neuronal plasticity15 and attenuation of HPA axis reactivity.10, 16 Despite several suggested modes for antidepressant action beyond monoamine elevation, the precise mechanisms at the cellular metabolism and pathway levels remain elusive. Activation of postsynaptic monoamine receptors in response to antidepressant treatment triggers intracellular signaling cascades relayed by G proteins that are coupled to several effector systems including adenylate cyclase, phospholipase C, phospholipase A2 and ion channels.17, 18 Second messengers like cyclic AMP and diacylglycerol induce intracellular protein phosphorylation events mediated by protein kinase A and protein kinase C, respectively. Phosphorylation events cause gene expression alterations through transcription factors like cyclic AMP response element-binding protein resulting in further downstream alterations.19, 20, 21, 22 Several studies have investigated the effects of antidepressants in unbiased transcriptomic or proteomic studies. Sillaber et al.23 identified a number of paroxetine-induced changes on the transcriptome of DBA/2 mice, including alterations in glial fibrillary acidic protein and brain-derived neurotrophic factor (BDNF) mRNA expression. Proteomic studies include paroxetine treatment effects on embryonic stem cell-derived neural cells24 and fluoxetine and venlafaxine effects on the rat hippocampus and frontal cortex25, 26 protein expression.
Metabolites reflect the ultimate response of an organism to any biological effect27 as they are the final products of interactions between gene expression, protein expression and the cellular environment.28 Therefore, metabolite profiling holds great promise for the identification of affected pathways in depression, and for the study of antidepressant drug treatment.
Recent studies have used metabolomic methods to interrogate psychiatric drug treatment effects. Atypical antipsychotic treatment influenced metabolism of specific lipid classes in patients with schizophrenia.29 Metabolomic studies on treatment effects of traditional Chinese medicine in rats identified potential biomarker candidates.30, 31 Ji et al.32 applied a pharmacometabolomic approach to guide targeted pharmacogenomic analyses in antidepressant responders versus non-responders. A pilot study in depressed patients of old age revealed alterations in plasma metabolite levels of GABA, glycerol and several fatty acids compared with controls.33 Most importantly, many of these alterations normalized after remission. Unfortunately, in this study, metabolomic changes could not be unambiguously attributed to depression or antidepressant treatment-related alterations.
In this study, we have for the first time analyzed metabolomic changes after chronic paroxetine treatment in DBA/2Ola mice. Using multivariate and univariate statistics, we have identified affected biochemical pathways downstream of serotonin reuptake inhibition and potential antidepressant drug targets in the hippocampus. Alterations in the plasma as the preferred specimen in a clinical setting represent a starting point for the implementation of a clinical biomarker assay for an early assessment of antidepressant treatment response.
Materials and methods
Antidepressant treatment of DBA/2Ola mice and organ sampling
Eight-week-old DBA/2OlaHsd mice were purchased from Harlan Laboratories (Harlan Winkelmann, Borchen, Germany). Upon arrival, mice were housed singly in standard cages and habituated for 2 weeks under standard laboratory conditions (food and water ad libitum, 12 h dark–light cycle: lights on 0700–1900 hours, 45–55% humidity, 21±2 °C). After 2 weeks, mice received either paroxetine at 10 mg kg−1 (1 mg paroxetine–hydrochloride–hemihydrate (Sigma-Aldrich, St Louis, MO, USA) in 1 ml tap water, thoroughly mixed before each application) or vehicle (tap water) twice per day (between 0800 and 0900 hours and 1800 and 1900 hours) by gavaging for 28 days. In the morning of day 29, mice received a final treatment, and 60 min later, the behavior in the forced swim test (FST), water temperature 25–26 °C, was observed during a 5-min test period. Immediately after the FST, mice were killed by an overdose of Forene isoflurane (Abbott, Wiesbaden, Germany), and blood was drawn by heart puncture and collected in EDTA tubes (Kabe Labortechnik, Nuembrecht-Elsenroth, Germany). Plasma was separated from serum by centrifugation (1300 g, 10 min) and immediately frozen in liquid nitrogen. Mice were perfused with 0.9% ice-cold saline (Merck, Darmstadt, Germany) and their brains sampled. Hippocampi were dissected, weighed, individually shock frozen in liquid nitrogen and stored at −80 °C until further analysis.
Metabolite analysis
Metabolite analysis was performed by the UC Davis Metabolomics Core Facility (http://www.metabolomics-core.ucdavis.edu/) using validated protocols and standard operation procedures (see Supplementary Figure S1). For quality control purposes, 30 standard metabolites were included in the samples in each randomized batch. FAMEs C08–C30 were used as internal references for retention index (RI) calibration, the Leco proprietary deconvolution protocol for peak finding (Leco, St Joseph, MI, USA) and the BinBase database (http://fiehnlab.ucdavis.edu/db/) for metabolite annotation.
Metabolite extraction
Per 10 mg of frozen hippocampus, 500 μl pre-chilled extraction solution (acetonitrile, isopropanole, water, 3:3:2 (v/v/v)) was added and the mixture was homogenized for 45 s using Tissue Master 125 (Omni International, Kennesaw, GA, USA). After centrifugation (13,000 g, 5 min), supernatants were dried and resuspended with 500 μl acetonitrile:water (1:1, v/v). After centrifugation (14 000 g, 2 min), supernatants were completely dried and stored in darkness under argon for further analysis. Plasma metabolite extraction was performed as described previously by Urayama et al.34
Sample derivatization and GC-time of flight-mass spectrometry metabolite profiling
Sample derivatization with methoxylamine hydrochloride in pyridine and N-methyl-N-trimethylsilyltrifluoroacetamide was performed as described previously.34 Gas chromatography (GC)-time of flight-mass spectrometry analysis was performed using an Agilent 6890N gas chromatograph (Agilent, Palo Alto, CA, USA) interfaced to a time-of-flight Pegasus III mass spectrometer (Leco). Automated injections were performed using an MPS2 programmable robotic multipurpose sampler (Gerstel, Mühlheim an der Ruhr, Germany). The GC system was fitted with both an Agilent injector and a Gerstel temperature-programmed injector, cooled injection system (model CIS 4), with a Peltier cooling source. An automated liner exchange (ALEX) designed by Gerstel was used to eliminate cross-contamination from sample matrix occurring between sample runs. Multiple baffled liners for the GC inlet were deactivated with 1-μl injections of MSTFA. The Agilent injector temperature was held constant at 250 °C, whereas the Gerstel injector was programmed (initial temperature 50 °C, hold 0.1 min and increased at a rate of 10 °C s−1 to a final temperature of 330 °C, hold time 10 min). Injections of 1 μl were made in the split (1:5) mode (purge time 120 s, purge flow 40 ml min−1).
Chromatography was performed on an Rtx-5Sil MS column (30 m × 0.25 mm i.d., 0.25-μm film thickness) with an Integra-Guard column (Restek, Bellefonte, PA, USA). Helium carrier gas was used at a constant flow of 1 ml min−1. The GC oven temperature program was 50 °C initial temperature, with 1-min hold time and ramping at 208 °C/min to a final temperature of 330 °C with 5-min hold time. Both the transfer line and the source temperatures were 250 °C. The Pegasus III time of flight-mass spectrometer ion source operated at −70 kV filament voltage. After a solvent delay of 350 s, mass spectra were acquired at 20 scans per second with a mass range of 50–500 m/z.
Data analysis and statistics
2-Monostearin, fructose, glucose, glucose-6-phosphate, glycine and phenylalanine were identified twice during chromatographic separation in different chemical derivatization states and their signal intensities were summed up for further analysis. Metabolite signal intensities were normalized by the total sum of all metabolite intensities. Pareto scaled metabolite signal intensities were then analyzed using MetaboAnalyst (http://www.metaboanalyst.ca).35
Downstream pathway analysis in the hippocampus and antidepressant drug targets
Identification of altered metabolites for subsequent pathway analysis in the hippocampus was performed by a combined univariate and multivariate data analysis strategy. Metabolites with altered concentrations were identified by SAM (significance analysis of microarrays)36 applying a q-value threshold of 15%. Multivariate data structure was revealed by partial least square—discriminant analysis considering metabolites with a variable importance in projection >1, using just one component (accuracy and R2>80%, Q2=0.57 and P<0.18 in permutation tests).37 By exclusively considering the overlap between the two statistically different methods, we improved robustness of data analysis and increased confidence in significantly altered metabolites.
Pathway analysis was performed using the Pathway Studio software v7.1 (Ariadne Genomics, Rockville, MD, USA), which contains literature-based relations between proteins, functional classes, small molecules and cellular processes. For an increased confidence, relations were only considered if there were at least two reported literature references.
For identification of affected molecular pathways, a search for common molecular upstream regulators was performed including proteins and functional classes and for downstream targets including proteins, functional classes and cellular processes of altered metabolites. A conservative hypergeometric test38 was performed to detect significant overrepresented (P<0.05) common regulators and targets. The background for each common regulator/target was set to the number of metabolites it relates to (minimum two references) in the Ariadne database. Owing to the conservative test, common regulators/targets with only a few interactions are highly penalized.38 Finally, metabolites along with their regulators and targets were grouped according to biological function into pathway clusters. In addition to the identified pathway clusters, potential antidepressant drug targets were identified. These comprise common regulators and targets (proteins and functional classes) that modulate these pathways.
Biomarker candidates in the hippocampus and plasma
To detect biomarker candidates for antidepressant treatment effects in the hippocampus, SAM was performed by applying a q-value threshold of q<0.05. Each metabolite is presented by a scatter plot indicating inter- and intra-group biological variabilities. Metabolite/behavior correlation was assessed using Pearson's correlation of metabolite intensities versus floating behavior in the FST. In addition, mice were grouped according to floating time independent of treatment group and corresponding average metabolite intensities are shown.
To detect biomarker candidates in the plasma, we correlated hippocampal and plasma metabolite intensities (SAMhippocampus q<0.05, Pearson's correlation P<0.10).
Results
We performed a comparative metabolomic study in chronic paroxetine- versus vehicle-treated DBA/2 mice. As expected, paroxetine treatment significantly reduced depression-like behavior in the FST, indicated by a reduced time of floating (P<0.0001, Figure 1a). Six biological replicates per group were selected for metabolite profiling based on immobility behavior in the FST (Figure 1b).
Behavioral analysis of paroxetine-treated mice. (a) Chronic paroxetine treatment reduced passive behavior in the forced swim test determined by time of floating, P<0.0001, tfloating (vehicle)=90.3±6.1 s (mean±s.e.m.), tfloating (paroxetine)=20.6±4.2 s. (b) Six biological replicates were selected for metabolomics analysis based on the time of floating, P=0.0014, tfloating (vehicle)=97.0±11.0 s (mean±s.e.m.), tfloating (paroxetine)=22.7±12.9 s.
A total of 270 metabolites were quantified in the hippocampus and plasma, of which 110 were of known identity including 25 sugars, 24 amino acids, 17 fatty acids, 12 lipids, 12 organic acids and 20 of other categories (Supplementary Tables S1 and S2).
Downstream pathway analysis and antidepressant drug target candidates in the hippocampus
For the investigation of affected hippocampal downstream pathways, we analyzed differences in metabolomic profiles using a combined univariate and multivariate data analysis strategy as described in the ‘Materials and methods’ section. In total, 28 metabolites including 9 amino acids, 5 sugars, 2 organic acids, 1 fatty acid, 4 of other categories and 7 of unknown identity were considered for pathway analysis (Table 1a). Twenty four significantly altered metabolites in the hippocampus were present at higher levels upon paroxetine compared with vehicle treatment. A log2 abundance factor distribution of all metabolites for the hippocampus and plasma is shown in Supplementary Figure S2, indicating slightly higher metabolite levels in paroxetine-treated hippocampi.
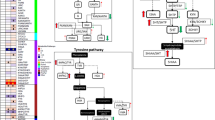
Identified pathway clusters included energy metabolism, amino-acid metabolism and hormone signaling (Figure 2). Regulators of these pathways, as illustrated in Figure 2 and summarized in Table 2, represent antidepressant drug targets that have the potential to modulate these pathways.
Pathway analysis of altered metabolites upon paroxetine treatment. Common targets and regulators of significant metabolites were identified by a conservative hypergeometric test expression analysis systematic explorer (EASE), score<0.05. Metabolites and targets/regulators were clustered into energy metabolism, amino acid metabolism and hormone signaling pathways. Green: metabolites, red: proteins, yellow: cellular processes, orange: functional classes.
Energy metabolism
Most profound alterations at the pathway level were found to be related to energy metabolism. Glucose, glucose-6-phosphate and fructose-6-phosphate, metabolites involved in glycolysis, were present at 2–4-fold higher levels upon paroxetine treatment. Levels of lactate, a metabolite linked to glycolysis, were reduced by 30%. Potential tricarboxylic acid (TCA) cycle alterations were reflected by a 2.5-fold increase in fumarate levels. Regulators of energy metabolism were identified as hexokinase 1 and 2, glucokinase, glycogen synthase 2, glucose transporter, fructose 1,6 diphosphatase, mannose phosphate isomerase and fructose bisphosphatase 1.
Amino acid metabolism
Eight proteinogenic amino acids were considered significant for pathway analysis (Ala, Ile, Leu, Pro, Ser, Thr, Tyr, Val), all being upregulated by paroxetine treatment (23–72% increase). All but two (Ser, Thr) amino acids are metabolically linked to TCA cycle intermediates linking amino-acid changes to energy metabolism alterations. Potential regulators are represented by 3-methyl-2-oxobutanoate dehydrogenase, an important enzyme in branched-chain amino acid (BCAA), Val, Leu, Ile, degradation and glutamate dehydrogenase, converting glutamate to alphaketoglutarate, a TCA cycle intermediate.
Hormone signaling
Hormone signaling was dominated by glucagon and insulin. Both control blood glucose levels and the pre-hormone angiotensinogen that is involved in blood pressure regulation.
Biomarker candidates in the hippocampus and plasma
Biomarker candidates were identified by univariate data analysis, as described in the ‘Materials and methods’ section. A total of 34 metabolites were found in the hippocampus (7 amino acids, 4 sugars, 2 fatty acids, 1 lipid, 2 organic acids, 1 of other categories and 17 of unknown identity), of which 24 were present at higher levels upon paroxetine treatment (Table 1b). Galactose-6-phosphate (4.88), fumaric acid (2.47), fructose (2.19) and xylulose (2.08) were among the metabolites with the greatest fold change and the most significant changes were found for threonic acid (q=0.009), xylulose (q=0.024) and valine (q=0.024). Supplementary Figure S3 provides detailed information on significant metabolites (scatter plots and correlation analysis). For many metabolites there was a high correlation between metabolite intensities and depression-like behavior in the FST. This represents a first hint that paroxetine-induced metabolite alterations may be linked to a depression-like phenotype and do not represent a mere side effect (Supplementary Figure S3).
For identification of plasma biomarker candidates, a correlation analysis of hippocampus versus plasma metabolite intensities was performed. Out of 17 hippocampus biomarker candidates (SAMhippocampus q<0.05) of known identity, 3 metabolites show high correlation values (two-tailed P<0.10) upon paroxetine treatment. Calculated Pearson's coefficient values were r=0.77 for GABA, r=0.75 for galactose-6-phosphate and r=−0.73 for leucine (Figure 3).
Discussion
Metabolomic studies hold great promise for the identification of molecular alterations upon drug treatment.29, 30, 31, 32, 39
To exclude any metabolite level alterations that are caused by environmental factors such as nutritional effects, animals with homogeneous genetic backgrounds and housed under controlled conditions are the preferred study objects. This way, inter-individual variability that is commonplace in patient studies can be avoided resulting in a better signal-to-noise ratio of the drug-elicited metabolite level changes. To our knowledge, this is the first study identifying metabolite signatures in chronically paroxetine-treated DBA/2 mice. We aimed at revealing treatment effects beyond elevation of serotonin levels in the synaptic cleft that are involved in therapeutic antidepressant effects. Understanding the cross-talk between altered metabolomic pathways will greatly enhance our understanding of the drug's mode of action and adverse side effects.
Affected downstream pathways in the hippocampus
The selective serotonin reuptake inhibitor paroxetine inhibits presynaptic serotonin transporters leading to enhanced serotonergic synaptic transmission, which was previously shown to be essential for therapeutic efficiency.6 Our results indicate that enhanced serotonin availability results in diverse downstream pathway alterations.
Most significantly, paroxetine treatment altered hippocampal energy metabolism (Figure 2, Tables 1a and 2), particularly the initial steps of glycolysis. Recent studies have linked antidepressant treatment to energy metabolism alterations. Scaini et al.40 demonstrated increased mitochondrial respiratory chain activity in selected brain regions after chronic antidepressant treatment. Prefrontal cortex, hippocampal and striatal, but not cerebellar citrate synthase and succinate dehydrogenase activities were increased after paroxetine treatment.41 Santos et al.42 found increased brain creatine kinase activity after chronic paroxetine treatment. All these findings indicate that in order to normalize metabolic hypoactivity, which is frequently observed in depressed patients, energy pathways are a pharmacological target worth pursuing. Increased ATP levels could also facilitate energy-dependent antidepressant-treatment-associated cellular processes like G-protein activity, protein kinase activities (protein kinase A, protein kinase C), synaptic activity (transport of ions and neurotransmitter uptake) and protein synthesis (BDNF).43
We further identified significant alterations in amino acid metabolism, which is closely linked to energy metabolism through TCA cycle intermediates. However, from our data it is not clear whether increased amino acid concentrations contribute to or are a consequence of altered energy metabolism and further studies are required to answer this question.
We also identified significantly altered neuroactive amino acid level including β-alanine, a glycine receptor agonist, and decreased levels of GABA, the most prominent inhibitory neurotransmitter, indicating a direct link of amino acid metabolism to neurotransmission. Furthermore, significantly increased levels of tyrosine, the amino acid precursor of epinephrine, could contribute to elevated noradrenergic neurotransmission, a target of selective noradrenaline reuptake inhibitors.
Biomarker candidates for antidepressant treatment effects in the hippocampus
Biomarkers for antidepressant treatment response that indicate expected treatment efficiency at an early time point are in great demand. The metabolites identified in this study can provide a basis for future in vivo studies using either nuclear magnetic resonance spectroscopy, or cerebrospinal fluid and blood samples, the latter being the preferred specimen for implementation of a clinical biomarker assay.
Branched chain amino acids
Concentrations of the essential amino acids valine, leucine and isoleucine increased by 50–70% upon paroxetine treatment. Elevated levels of these amino acids, especially leucine, have been shown to increase protein synthesis through increased mammalian target of rapamycin signaling in liver, skeletal muscle, kidney and adipose tissues.44, 45, 46 Until now, however, there is no evidence that BCAAs also induce protein synthesis in the brain. Mammalian target of rapamycin signaling, which is closely linked to synaptic plasticity,47 was shown to be inhibited by the selective serotonin reuptake inhibitor sertraline leading to decreased, not elevated, protein synthesis,48 seen in other tissues. Rapamycin, an inhibitor of mammalian target of rapamycin signaling exhibited antidepressant-like effects in mice and rats,49 whereas antidepressant effects of ketamine in treatment-resistant depressed patients were attributed to activated mammalian target of rapamycin signaling.50
Different pathways could be affected by increased BCAA levels in the hippocampus since BCAA metabolism is directly connected to energy metabolism. Specifically, oxidative BCAA degradation leads to Krebs cycle intermediates.51, 52 Thus, alterations in energy metabolism upon paroxetine treatment could, at least in part, be influenced by altered BCAA concentrations. Furthermore, alterations in Krebs cycle intermediates were shown to result in altered neurotransmitter synthesis,53 suggesting that increased BCAA levels could also influence synaptic transmission.
Biomarker candidates for antidepressant treatment effects in the plasma
Metabolite alterations in the plasma were less pronounced than those in the hippocampus. This finding is not unexpected as paroxetine primarily targets cerebral neurotransmission. To see whether central nervous system alterations are also reflected in the plasma, we performed a brain/plasma metabolite correlation analysis. We identified GABA, galactose-6-phosphate and leucine as biomarker candidates for antidepressant treatment effects. All three plasma biomarker candidates represent alterations also observed in the brain. Further studies need to validate these findings in humans by a targeted analysis in specimens from antidepressant treatment responders versus non-responders.
Future studies
Although we have identified a significant intensity level/phenotype correlation for a number of metabolites (Supplementary Figure S3), we cannot unambiguously associate identified pathway alterations or biomarker candidates with antidepressant therapeutic effects. To address this issue further, one line of investigation could include a pharmacological analysis of different mouse models of depression including chronic mild stress,54 social defeat55, 56 or early-life stress through maternal separation,57 wherein antidepressant treatment effects could be more directly associated with decreased depression-like behavior. Directly targeting the identified pathways in mouse models of depression may also help in this endeavor. In this pilot study, we have identified molecular changes upon antidepressant treatment at the metabolome level in DBA/2 mice that reflect pathway alterations beyond monoamine reuptake inhibition. On the basis of pathway information, we revealed putative antidepressant drug targets and biomarker candidates for the assessment of antidepressant treatment effects elicited through novel modes of action.
References
Burmeister M, McInnis MG, Zollner S . Psychiatric genetics: progress amid controversy. Nat Rev Genet 2008; 9: 527–540.
World Health Organization. The World Health Report 2001 – Mental Health: New Understanding, New Hope; 2001.
Anderson IM . Selective serotonin reuptake inhibitors versus tricyclic antidepressants: a meta-analysis of efficacy and tolerability. J Affect Disord 2000; 58: 19–36.
Berton O, Nestler EJ . New approaches to antidepressant drug discovery: beyond monoamines. Nat Rev Neurosci 2006; 7: 137–151.
Wong ML, Licinio J . From monoamines to genomic targets: a paradigm shift for drug discovery in depression. Nat Rev Drug Discov 2004; 3: 136–151.
Delgado PL . How antidepressants help depression: mechanisms of action and clinical response. J Clin Psychiatry 2004; 65 (Suppl 4): 25–30.
Delgado PL . Depression: the case for a monoamine deficiency. J Clin Psychiatry 2000; 61 (Suppl 6): 7–11.
Ruhe HG, Mason NS, Schene AH . Mood is indirectly related to serotonin, norepinephrine and dopamine levels in humans: a meta-analysis of monoamine depletion studies. Mol Psychiatry 2007; 12: 331–359.
Nickel T, Sonntag A, Schill J, Zobel AW, Ackl N, Brunnauer A et al. Clinical and neurobiological effects of tianeptine and paroxetine in major depression. J Clin Psychopharmacol 2003; 23: 155–168.
Holsboer F . Stress, hypercortisolism and corticosteroid receptors in depression: implications for therapy. J Affect Disord 2001; 62: 77–91.
Michelson D, Galliven E, Hill L, Demitrack M, Chrousos G, Gold P . Chronic imipramine is associated with diminished hypothalamic-pituitary-adrenal axis responsivity in healthy humans. J Clin Endocrinol Metab 1997; 82: 2601–2606.
Brady LS, Gold PW, Herkenham M, Lynn AB, Whitfield Jr HJ . The antidepressants fluoxetine, idazoxan and phenelzine alter corticotropin-releasing hormone and tyrosine hydroxylase mRNA levels in rat brain: therapeutic implications. Brain Res 1992; 572: 117–125.
Malberg JE, Eisch AJ, Nestler EJ, Duman RS . Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci 2000; 20: 9104–9110.
Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 2003; 301: 805–809.
Vaidya VA, Duman RS . Depression –emerging insights from neurobiology. Br Med Bull 2001; 57: 61–79.
Binder EB, Nemeroff CB . The CRF system, stress, depression and anxiety-insights from human genetic studies. Mol Psychiatry 2010; 15: 574–588.
Gilman AG . G proteins in signal transduction: transducers of receptor-generated signals. Annu Rev Biochem 1987; 56: 615–649.
Birnbaumer L . G proteins in signal transduction. Annu Rev Pharmacol Toxicol 1990; 30: 675–705.
Gould TD, Manji HK . Signaling networks in the pathophysiology and treatment of mood disorders. J Psychosom Res 2002; 53: 687–697.
Schwaninger M, Schofl C, Blume R, Rossig L, Knepel W . Inhibition by antidepressant drugs of cyclic AMP response element-binding protein/cyclic AMP response element-directed gene transcription. Mol Pharmacol 1995; 47: 1112–1118.
Shelton RC . Intracellular mechanisms of antidepressant drug action. Harv Rev Psychiatry 2000; 8: 161–174.
Sun MK, Alkon DL . Protein kinase C substrate activators: potential as novel antidepressants. Drug Dev Res 2005; 65: 77–91.
Sillaber I, Panhuysen M, Henniger MS, Ohl F, Kuhne C, Putz B et al. Profiling of behavioral changes and hippocampal gene expression in mice chronically treated with the SSRI paroxetine. Psychopharmacology (Berl) 2008; 200: 557–572.
McHugh PC, Rogers GR, Loudon B, Glubb DM, Joyce PR, Kennedy MA . Proteomic analysis of embryonic stem cell-derived neural cells exposed to the antidepressant paroxetine. J Neurosci Res 2008; 86: 306–316.
Carboni L, Vighini M, Piubelli C, Castelletti L, Milli A, Domenici E . Proteomic analysis of rat hippocampus and frontal cortex after chronic treatment with fluoxetine or putative novel antidepressants: CRF1 and NK1 receptor antagonists. Eur Neuropsychopharmacol 2006; 16: 521–537.
Khawaja X, Xu J, Liang JJ, Barrett JE . Proteomic analysis of protein changes developing in rat hippocampus after chronic antidepressant treatment: implications for depressive disorders and future therapies. J Neurosci Res 2004; 75: 451–460.
Fiehn O . Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol 2002; 48: 155–171.
Kaddurah-Daouk R, Krishnan KR . Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology 2009; 34: 173–186.
Kaddurah-Daouk R, McEvoy J, Baillie RA, Lee D, Yao JK, Doraiswamy PM et al. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Mol Psychiatry 2007; 12: 934–945.
Su ZH, Li SQ, Zou GA, Yu CY, Sun YG, Zhang HW et al. Urinary metabonomics study of anti-depressive effect of Chaihu-Shu-Gan-San on an experimental model of depression induced by chronic variable stress in rats. J Pharm Biomed Anal 2011; 55: 533–539.
Dai Y, Li Z, Xue L, Dou C, Zhou Y, Zhang Y et al. Metabolomics study on the anti-depression effect of xiaoyaosan on rat model of chronic unpredictable mild stress. J Ethnopharmacol 2010; 128: 482–489.
Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther 2011; 89: 97–104.
Paige LA, Mitchell MW, Krishnan KR, Kaddurah-Daouk R, Steffens DC . A preliminary metabolomic analysis of older adults with and without depression. Int J Geriatr Psychiatry 2007; 22: 418–423.
Urayama S, Zou W, Brooks K, Tolstikov V . Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Commun Mass Spectrom 2010; 24: 613–620.
Xia J, Psychogios N, Young N, Wishart DS . MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 2009; 37 (Web Server issue): W652–W660.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121.
Wold S, Sjöström M, Eriksson L . PLS-regression: a basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems 2001; 58: 109–130.
Hosack DA, Dennis Jr G, Sherman BT, Lane HC, Lempicki RA . Identifying biological themes within lists of genes with EASE. Genome Biol 2003; 4: R70.
Watkins SM, German JB . Metabolomics and biochemical profiling in drug discovery and development. Curr Opin Mol Ther 2002; 4: 224–228.
Scaini G, Maggi DD, De-Nes BT, Goncalves CL, Ferreira GK, Teodorak BP et al. Activity of mitochondrial respiratory chain is increased by chronic administration of antidepressants. Acta Neuropsychchiatrica 2011; 23: 112–118.
Scaini G, Santos PM, Benedet J, Rochi N, Gomes LM, Borges LS et al. Evaluation of Krebs cycle enzymes in the brain of rats after chronic administration of antidepressants. Brain Res Bull 2010; 82: 224–227.
Santos PM, Scaini G, Rezin GT, Benedet J, Rochi N, Jeremias GC et al. Brain creatine kinase activity is increased by chronic administration of paroxetine. Brain Res Bull 2009; 80: 327–330.
Moretti A, Gorini A, Villa RF . Affective disorders, antidepressant drugs and brain metabolism. Mol Psychiatry 2003; 9: 773–785.
Lynch CJ, Hutson SM, Patson BJ, Vaval A, Vary TC . Tissue-specific effects of chronic dietary leucine and norleucine supplementation on protein synthesis in rats. Am J Physiol Endocrinol Metab 2002; 283: E824–E835.
Lynch CJ, Patson BJ, Anthony J, Vaval A, Jefferson LS, Vary TC . Leucine is a direct-acting nutrient signal that regulates protein synthesis in adipose tissue. Am J Physiol Endocrinol Metab 2002; 283: E503–E513.
Nishitani S, Ijichi C, Takehana K, Fujitani S, Sonaka I . Pharmacological activities of branched-chain amino acids: specificity of tissue and signal transduction. Biochem Biophys Res Commun 2004; 313: 387–389.
Hoeffer CA, Klann E . mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci 2010; 33: 67–75.
Lin CJ, Robert F, Sukarieh R, Michnick S, Pelletier J . The antidepressant sertraline inhibits translation initiation by curtailing mammalian target of rapamycin signaling. Cancer Res 2010; 70: 3199–3208.
Cleary C, Linde JA, Hiscock KM, Hadas I, Belmaker RH, Agam G et al. Antidepressive-like effects of rapamycin in animal models: Implications for mTOR inhibition as a new target for treatment of affective disorders. Brain Res Bull 2008; 76: 469–473.
Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 2010; 329: 959–964.
Murin R, Mohammadi G, Leibfritz D, Hamprecht B . Glial metabolism of valine. Neurochem Res 2009; 34: 1195–1203.
Murin R, Mohammadi G, Leibfritz D, Hamprecht B . Glial metabolism of isoleucine. Neurochem Res 2009; 34: 194–204.
Aquilani R, Boselli M, Boschi F, Viglio S, Iadarola P, Dossena M et al. Branched-chain amino acids may improve recovery from a vegetative or minimally conscious state in patients with traumatic brain injury: a pilot study. Arch Phys Med Rehabil 2008; 89: 1642–1647.
Willner P . Chronic mild stress (CMS) revisited: consistency and behavioural-neurobiological concordance in the effects of CMS. Neuropsychobiology 2005; 52: 90–110.
Kudryavtseva NN, Bakshtanovskaya IV, Koryakina LA . Social model of depression in mice of C57BL/6J strain. Pharmacol Biochem Behav 1991; 38: 315–320.
Rygula R, Abumaria N, Flügge G, Fuchs E, Rüther E, Havemann-Reinecke U . Anhedonia and motivational deficits in rats: impact of chronic social stress. Behav Brain Res 2005; 162: 127–134.
Millstein RA, Holmes A . Effects of repeated maternal separation on anxiety- and depression-related phenotypes in different mouse strains. Neurosci Biobehav Rev 2007; 31: 3–17.
Acknowledgements
This work was supported by a BMBF QuantPro Grant and the Max Planck Society. CW was supported by the International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS-LS) and the Graduate School of Systemic Neurosciences (GSN-LMU).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Webhofer, C., Gormanns, P., Tolstikov, V. et al. Metabolite profiling of antidepressant drug action reveals novel drug targets beyond monoamine elevation. Transl Psychiatry 1, e58 (2011). https://doi.org/10.1038/tp.2011.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2011.56
Keywords
This article is cited by
-
Elevated O-GlcNAcylation induces an antidepressant-like phenotype and decreased inhibitory transmission in medial prefrontal cortex
Scientific Reports (2020)
-
Metabolomic analysis of animal models of depression
Metabolic Brain Disease (2020)
-
A comprehensive metabolomics investigation of hippocampus, serum, and feces affected by chronic fluoxetine treatment using the chronic unpredictable mild stress mouse model of depression
Scientific Reports (2019)
-
Ketamine’s antidepressant effect is mediated by energy metabolism and antioxidant defense system
Scientific Reports (2017)
-
Delineation of molecular pathway activities of the chronic antidepressant treatment response suggests important roles for glutamatergic and ubiquitin–proteasome systems
Translational Psychiatry (2017)