Abstract
Hostility is a multidimensional personality trait with changing expression over the life course. We performed a genome-wide association study (GWAS) of the components of hostility in a population-based sample of Finnish men and women for whom a total of 2.5 million single-nucleotide polymorphisms (SNPs) were available through direct or in silico genotyping. Hostility dimensions (anger, cynicism and paranoia) were assessed at four time points over a 15-year interval (age range 15–30 years at phase 1 and 30–45 years at phase 4) in 982–1780 participants depending on the hostility measure. Few promising areas from chromosome 14 at 99 cM (top SNPs rs3783337, rs7158754, rs3783332, rs2181102, rs7159195, rs11160570, rs941898, P values <3.9 × 10−8 with nearest gene Enah/Vasp-like (EVL)) were found suggestively to be related to paranoia and from chromosome 7 at 86 cM (top SNPs rs802047, rs802028, rs802030, rs802026, rs802036, rs802025, rs802024, rs802032, rs802049, rs802051, P values <6.9 × 10−7 with nearest gene CROT (carnitine O-octanoyltransferase)) to cynicism, respectively. Some shared suggestive genetic influence for both paranoia and cynicism was also found from chromosome 17 at 2.8 cM (SNPs rs12936442, rs894664, rs6502671, rs7216028) and chromosome 22 at 43 cM (SNPs rs7510759, rs7510924, rs7290560), with nearest genes RAP1 GTPase activating protein 2 (RAP1GAP2) and KIAA1644, respectively. These suggestive associations did not replicate across all measurement times, which warrants further study on these SNPs in other populations.
Similar content being viewed by others
Introduction
Hostility is a personality trait characterizing how trustworthy individuals perceive other people and how they handle these feelings toward others. The cognitive component of hostility characterizes cynical and distrustful attitudes, which is the primary reference of the term hostility,1 whereas the affective component reflects feelings of irritability and anger. The behavioral component covers expression of these attitudes and feelings as either expressing them out, that is, aggression, or as suppressing or repressing them. Hostility traits have been found to be related to various social and health problems, such as criminality and violence,2, 3 isolation and relationship aggression,4 depression,5 cardiovascular diseases6 and all-cause mortality risk,7 although the findings are not entirely consistent.8 Identifying the origins of hostility may help to understand the developmental paths related to hostility and to develop effective preventions to reduce problems related to hostile behaviors.
Both genetic and environmental factors are involved in the development of hostility,9 with heritability estimated to be ∼30–50%.10, 11, 12 However, the molecular nature of the genetic background and the specific regions of the genome that underlie hostility remain mainly unknown. To our knowledge, only one genome scan study of hostility has been published to date.13 That study covered 387 autosomal short-tandem-repeat polymorphisms and did not find significant linkage with hostility.13 In the present study, we report a large-scale genome-wide association study (GWAS) of hostility where over 2.5 million single-nucleotide polymorphisms (SNPs) were analyzed, thereby mapping the most potentially significant areas of the genome regarding hostility for further inspection and providing preliminary evidence of the genetic basis of hostility.
As cognitive, affective and behavioral components of hostility may vary in their etiology and have different genetic backgrounds,14 we used three different scales of hostility, each of which was measured four times over a 15-year time span extending from adolescence and young adulthood (age 15–30 years) into adulthood (age 30–45 years) in a Caucasian Finnish population. It has been argued that personality is still transient and amenable to environmental effects in young adulthood, but between 30 and 50 years of age, it begins to stabilize and genetic effects become more prominent.11 Thus, an additional aim of the present study was to test whether genetic effects underlying hostility are stable across different ages or whether they gain importance with advanced age in adulthood.
Materials and methods
Population and study design
Participants were from the population-based prospective Young Finns (YF) cohort study, which started in 1980 with 3596 boys and girls from different geographical areas of Finland.15 The genome of the participants was genotyped in 2009 and personality tests assessing hostility were administered in four follow-up phases in 1992, 1997, 2001 and 2007. At the baseline of the present study (1992), participants were 15, 18, 21, 24, 27 and 30 years old, and they were followed up for 15 years until they were 30, 33, 36, 39, 42 and 45 years, respectively. The final study sample with complete measurements consisted of 982–1781 men and women depending on the measure of hostility (n for anger between the four measurement phases ranges between 1619 and 1776, n for cynicism between 1622 and 1781 and n for paranoia between 1622 and 1780).
Measures of hostility components
We assessed three aspects of hostility, that is, cynicism and paranoia, which both reflect the cognitive component, and anger, which represents the affective component. Cynicism was measured with a seven-item self-completion cynicism scale derived from the Minnesota Multiphasic Personality Inventory (for example, ‘It is safer to trust nobody’).16, 17 Paranoia was assessed with the six-item self-completion paranoid ideation subscale of the Symptom Checklist-90R (for example, ‘Others do not give me proper credit for my achievements’).18 Anger was assessed with a seven-item Irritability Scale of the Buss-Durkee Hostility Inventory (for example, ‘I lose my temper easily but get over it quickly’).19 Detailed description of the scales has been published in previous papers.9, 20 Response format for all scales was on a five-point scale, ranging from totally disagree (1) to totally agree (5), and the mean of each scale was calculated for only those who had responded to at least 50% of the items on the scale. In addition, for each scale we calculated the mean score over the four measurements in 1992, 1997, 2001 and 2007 to capture a more stable trait of hostility. Reliability for the four-measurement mean score was high, with Cronbach's α being 0.85, 0.84 and 0.82 for cynicism, paranoia and anger scales, respectively.
Genotyping and quality control of YF study
The genome-wide SNP genotyping of YF study was done by a custom Illumina BeadChip (San Diego, CA, USA) containing 670 000 SNPs and copy-number variant probes from 2442 YF participants (1123 males and 1319 females). The custom 670K chip shares 562 643 SNPs in common with the Illumina Human610 BeadChip. Genotypes were called using Illumina's clustering algorithm (Illuminus).21 A total of 2556 samples were genotyped. After initial clustering, we removed 2 subjects for poor call rates (call rate <0.90), and 54 samples failed subsequent quality control (that is, duplicated samples, heterozygosity, low call rate or custom SNP fingerprint genotype discrepancy). The following filters were applied to the remaining data: minor allele frequency 0.01, genotyped call rate (GENO) 0.05, MIND 0.05 and Hardy–Weinberg equilibrium 1 × 10−6. Of 2500 individuals, 3 were removed for low genotyping (MIND >0.05), 11 766 markers were excluded based on Hardy–Weinberg equilibrium test (P⩽1 × 10−6), 7746 SNPs failed missingness test (GENO >0.05), 34 596 SNPs failed frequency test (minor allele frequency <0.01) and 1 individual failed gender check. None were removed by subsequent heterozygosity check. In that point, there were 546 770 SNPs and 2496 individuals who were utilized to generate an identity-by-descent matrix file in PLINK.22 There were 51 pairs of individuals with pi-hat >0.2, and thus these individuals were removed because of possible relatedness. One of the pair was removed using greater missingness as criteria. After final frequency and genotyping running, there was 546 677 SNPs available from a sample of 2442 YF subjects. Genotype imputation was performed for the YF SNP data using MACH23 with the HapMap (phase II, release 22 CEU, NCBI build 36, dbSNP 126) haplotypes as reference.
Statistics of GWAS
Quasi-continuous mean variables of hostility subscales were Box–Cox transformed. Residuals were obtained using linear regression model in which hostility variables were adjusted for sex and age in order to control the most obvious environmental factors related to hostility. Residuals were standardized (mean 0, s.d. 1) and their distributions were confirmed to be very close to normal by visual Q-Q plot analysis. We also verified that the estimates for the β-coefficients from the GWAS are not driven by few outliers by plotting leverage vs standardized residuals plots for the residuals.
We have an 80% power of identifying SNPs that explain at least 4% of the variability with sample size of 985 (mean of four measurement). For the four measurements within each hostility scale the sample sizes are increased to ∼1780 subjects. These analyses were powered to detect the effects of common variants down to 2.1% of explained variability.
Tests for additive genetic effects were carried out on a linear scale using linear regression. Genotypes were coded as 0, 1 or 2 when the SNP was genotyped and by dosage (scale 0–2) when imputed. These tests were performed to assess association of SNPs with the standardized residuals using PLINK22 for the genotyped data. ProbABEL24 was used to fit the model, taking account of the genotype uncertainty at imputed SNPs. The P-values were combined from the analysis by favoring genotyped tests over imputed ones. The Q-Q and Manhattan plots were drawn for the analysis of the results. The P-value for genome-wide significance was set at P<9 × 10−8, corresponding to a target α of 0.05 with a Bonferroni correction for 550 000 million independent tests with direct genotyping. Cynicism was normally distributed, whereas the distributions of paranoia and anger were slightly positively skewed. Thus Box–Cox transformations were used for all the outcomes.
Results
As shown in Table 1, the average age of the genotyped sample is 37.56 (s.d.=5.03). The bivariate correlations between hostility measures are shown in Table 2. The stability of the measures (r's range 0.45–0.69) as well as their bivariate correlations (r's range 0.38–0.77) are moderate (all P's <0.001). Cynicism and paranoia correlate higher with each other than with anger. Younger participants scored higher on the three hostility measures (r=−0.12, P<0.001, r=−0.08, P=0.01, and r=−0.05, P=0.123 for mean cynicism, paranoia and anger, respectively). Females scored higher on anger (r=−0.20, P<0.001) and males on cynicism (r=0.18, P<0.001) and paranoia (r=0.09, P<0.01). All the subsequent models were therefore adjusted for sex and age.
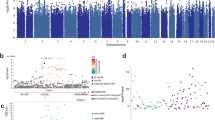
We tested 2 577 640 SNPs for association with the three hostility scales measured in four different time points. The top SNPs derived from SNPs with P-values ⩽1 × 10−5 are presented in the Tables 3 and 4. Table 3 shows the top SNPs when hostility is measured as a mean score of four measurement phases ((phase 1+phase 2+phase 3+phase 4/4)). Chromosome 14 at 99 cM (SNPs rs3783337, rs7158754, rs3783332, rs2181102, rs7159195, rs11160570, rs941898) predicted suggestively the mean paranoia during the 15 years at the genome-wide statistical significance (P<9 × 108, Table 2). However, this suggestive association did not replicate at each single measurement point over time (Table 4). Table 4 shows the top SNPs when the most significant associations are selected, irrespective of measurement phase (selected from phase 1, phase 2, phase 3 or phase 4). The most significant SNP suggestively associated with anger was found on chromosome 17 at 11 cM SNP rs11656526 (P-value <9 × 10−8, Table 4) for anger measured in 1992. Also, loci on chromosome 6 at 6.7 cM seemed promising when predicting anger in 2007, which shows the most reliable results for anger according to Q-Q plot analyses. However, these suggestive associations did not replicate in other measurement years, and hence the stability of these associations was weak.
Table 5 shows those SNPs that replicate at different measurement phases or at different scales. The most systematic replicating evidence for suggestive genetic effects was found for cynicism, although the significance levels (P<1 × 105) did not reach the Bonferroni corrected genome-wide significance level. Promising SNPs suggestively predicting cynicism were found on chromosome 7 at 86 cM (rs802047, rs802028, rs802030, rs802026, rs802036, rs802025, rs802024, rs802032, rs802049, rs802051), which replicated on two different measurements of cynicism (1992 and 1997) as well as the first four of the SNPs on above on the mean of all four measurements of cynicism (Table 5). In addition, SNPs in chromosome 22 at 43 cM (rs7510759, rs7510924) were associated with cynicism in 1997 and the mean of all four measurements of cynicism. The genetic background of different components of hostility appears to be largely distinct from each other, although a group of SNPs from chromosome 17 at 2.8 cM (rs12936442, rs894664, rs6502671, rs7216028) and from chromosome 22 at 43 cM (rs7510759, rs7510924, rs7290560) and at 36 cM (rs8136107) were suggestively associated with both cynicism and paranoia. Replications of the genetic linkage between different measurement of hostility and different measurement years are presented in Table 5.
Discussion
Our study reports results of a large-scale GWA analysis of hostility, with hostility measured in four follow-ups across 15 years of time span with three different scales. Although only few associations achieved genome-wide significance, many associations approached significance. We attempted to capture more reliable findings of the genotype over time by using the mean of hostility levels in the four time points as the outcome. Most of the suggestive associations did not replicate across measurement times, which undermines the robustness of the single significant associations. These suggestive associations should therefore be interpreted with appropriate caution. The inconsistent findings resemble those from many previous GWA studies of personality, most of which have not yet found robust evidence for specific candidate genes.13, 25, 26, 27
The strongest associations were found for mean score of paranoia with a number of closely linked SNPs in chromosome 14 at 99 cM, although this suggestive association had limited replicability over time. Chromosome 14 at∼100 cM has been previously linked to neuroticism and anxiety27, 28 and at 103 cM to bipolar disorder.29 The present study thus adds evidence that this region may include genetic markers or determinants for general anxiety and distrust (that is, paranoia) as well as for susceptibility to psychiatric diagnoses involving distrust against others. The finding that the mean paranoia for four different time points had significant genetic linkage, but single measurements of paranoia did not, may imply that paranoia as a stable trait has wider genetic basis, but high distrust in one point in time may depend more on transient environmental factors and be more prone to fluctuate. The closest gene for the found paranoia linked SNPs is EVL gene in chromosome 14, which is proposed as a possible candidate gene for colorectal cancer.30
Another significant genomic region found in the current study is in chromosome 17 at 2.8 cM, which was suggestively linked with both paranoia and cynicism in the most recent measurement when the participants were at age 30–45 years. The closest gene for this region is RAP1GAP2, which affects GTPase-activating protein, has a role in regulating the platelet aggregation, and is expressed especially in heart, testis and blood leukocytes, and also in stomach, pancreas and intestines, and slightly in brain.31 Thus, this might also be a possible link between hostility and health problems. Both cynicism and paranoia mean scores were also associated with a region in chromosome 22 at 36 cM and at 43 cM for which the closest gene is KIAA1644. Neither RAP1GAP2 nor KIAA1644 have previously been linked to personality traits, although chromosome 22 at 36 cM has been linked to bipolar disorder and schizophrenia.32
Although the results for cynicism did not reach the Bonferroni corrected statistical significance level, there were many marginally significant associations. Especially, areas on chromosome 7 at 86 cM were related to cynicism in 1992, 1997 and mean of cynicism measurements. The nearest gene for this region is CROT (carnitine O-octanoyltransferase) that affects fatty acid functioning in a cell level and is expressed at least in mice almost everywhere in the body, especially in liver and intestines, and also slightly in heart and brain.33
The observed suggestive associations may have some theoretical implications. Theoretically, cynicism is assumed to develop more in response to environmental experiences, which may explain the less significant relation to genetic background. However, it may be that there are multiple overlapping genetic effects and gene × gene and gene × environment interactions that prevent SNPs to reach the Bonferroni corrected significance level. Same locations in chromosome 17 at 2.8 cM and chromosome 22 at 36 and 43 cM were associated with both cynicism and paranoia, which may imply shared genetic background with these hostility dimensions. Such hostile attitudes might be seen as core of the hostility construct.1, 7, 9 Anger, on the other hand, is theoretically a separate construct having its developmental roots in temperament-like characteristics.34 Our finding that anger did not share similar genetic background with cynicism or paranoia implies that the consideration of anger as a separate construct seems justified also from the genetic perspective. The phenotypic and genotypic differences behind hostility measures may thus in part explain the mixed findings between hostility and cardiovascular health.8
Measuring complex personality traits, like hostility, involves challenges of accurate measurement of the phenotype. Measurement error due to imperfect assessment of the phenotype reduces the ability to capture stabile phenotype over time (test–retest correlations) and introduces time-specific variance in the measures. The lack of adjustment for relevant environmental factors influencing personality development may partly explain why the GWAS findings of personality traits rarely replicate in different time points or in different samples. This is not a unique problem to our study, as previous studies with well-established personality scales, for example, Temperament and Character Inventory25, 35 and ‘Big Five’,26, 36, 37 have rarely found consistent associations with GWAS.
In summary, this GWAS showed preliminary evidence for specific regions possibly related to hostility. The suggestive associations were small in magnitude and did not replicate across all measurement times, and thus they warrant further study in other populations. Single SNPs are likely to have small and thereby variable effects on personality traits, and many real effects may be lost in plenty of associations because of insufficient statistical power and measurement imprecision related to the identification of the phenotype. Accumulating evidence from several cohorts should provide more accurate and reliable data on the genetic background of hostility and other personality traits.
References
Greenglass ER, Julkunen J . Construct validity and sex differences in Cook-Medley hostility. Person Individ Diff 1989; 10: 209–218.
Soyka M, Graz C, Bottlender R, Dirschedl P, Schoech H . Clinical correlates of later violence and criminal offences in schizophrenia. Schizophr Res 2007; 94: 89–98.
Abu-Akel A, Abushua’leh K . ‘Theory of mind’ in violent and nonviolent patients with paranoid schizophrenia. Schizophr Res 2004; 69: 46–53.
Vandervoort D . Quality of social support in mental and physical health. Curr Psychol 1999; 18: 205–222.
Heponiemi T, Elovainio M, Kivimäki M, Pulkki L, Puttonen S, Keltikangas-Järvinen L . The longitudinal effects of social support and hostility on depressive tendencies. Soc Sci Med 2006; 63: 1374–1382.
Chida Y, Steptoe A . The association of anger and hostility with future coronary heart disease: a meta-analytic review of prospective evidence. J Am Coll Cardiol 2009; 53: 936–946.
Miller TQ, Smith TW, Turner CW, Guijarro ML, Hallet AJ . A meta-analytic review of research on hostility and physical health. Psychol Bull 1996; 119: 322–348.
Rozanski A, Blumenthal JA, Kaplan J . Impact of psychological factors on the pathogenesis of cardiovascular disease and implications for therapy. Circulation 1999; 99: 2192–2217.
Merjonen P, Pulkki-Råback L, Lipsanen J, Lehtimäki T, Rontu R, Viikari J et al. Development of adulthood hostile attitudes: childhood environment and serotonin receptor gene interactions. Pers Relationship 2011; 18: 184–197.
Hur YM . Nonadditive genetic effects on hostility in South Korean adolescent and young adult twins. Twin Res Hum Genet 2006; 9: 637–641.
Rebollo I, Boomsma DI . Genetic analysis of anger: genetic dominance or competitive sibling interaction. Behav Genet 2006; 36: 216–228.
Coccaro EF, Bergeman CS, Kavoussi RJ, Seroczynski AD . Heritability of aggression and irritability: a twin study of the Buss-Durkee aggression scales in adult male subjects. Biol Psychiatry 1997; 41: 273–284.
Knox SS, Wilk JB, Zhang Y, Weidner G, Ellison RC . A genome scan for hostility: the National Heart, Lung, and Blood Institute Family Heart Study. Mol Psychiatry 2004; 9: 124–126.
Gustavsson JP, Pedersen NL, Asberg M, Schalling D . Exploration into the sources of individual differences in aggression-, hostility- and anger-related (AHA) personality traits. Pers Individ Diff 1996; 21: 1067–1071.
Raitakari OT, Juonala M, Rönnemaa T, Keltikangas-Järvinen L, Räsänen L, Pietikäinen M et al. Cohort profile: the Cardiovascular Risk in Young Finns Study. Int J Epidemiol 2008; 37: 1220–1226.
Comrey AL . A factor analysis of items on the MMPI paranoia scale. Educ Psychol Meas 1958; 18: 99–107.
Comrey AL . A factor analysis of items on the MMPI hysteria scale. Educ Psychol Meas 1957; 17: 586–592.
Derogatis LR, Cleary PA . Confirmation of the dimensional structure of the SCL-90: a study in construct validation. J Clin Psychol 1977; 33: 981–989.
Buss AH, Durkee A . An inventory for assessing different kinds of hostility. J Consult Psychol 1957; 21: 343–349.
Merjonen P, Pulkki-Råback L, Puttonen S, Keskivaara P, Juonala M, Telama R et al. Anger is associated with subclinical atherosclerosis in low SES but not in higher SES men and women. The Cardiovascular Risk in Young Finns Study. J Behav Med 2008; 31: 35–44.
Teo YY, Inouye M, Small KS, Gwilliam R, Deloukas P, Kwiatkowski DP et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics 2007; 23: 2741–2746.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Li Y, Abecasis GR . Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet 2006; S79: 2290.
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM . GenABEL: an R library for genome-wide association analysis. Bioinformatics 2007; 23: 1294–1296.
Verweij KJH, Zietsch BP, Medland SE, Gordon SD, Benyamin B, Nyholt DR et al. A genome-wide association study of Cloninger's temperament scales: implications for the evolutionary genetics of personality. Biol Psychol 2010; 85: 306–317.
Terracciano A, Sanna S, Uda M, Deiana B, Usala G, Busonero F et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry 2010; 15: 647–656.
Wray NR, Middeldorp CM, Birley AJ, Gordon SD, Sullivan PF, Visscher PM et al. Genome-wide linkage analysis of multiple measures of neuroticism of 2 large cohorts from Australia and the Netherlands. Arch Gen Psychiatry 2008; 65: 649–658.
Middeldorp CM, Hottenga JJ, Slagboom PE, Sullivan PF, de Geus EJ, Posthuma D et al. Linkage on chromosome 14 in a genome-wide linkage study of a broad anxiety phenotype. Mol Psychiatry 2008; 13: 84–89.
Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–678.
Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006; 314: 268–274.
Schultess J, Danielewski O, Smolenski AP . Rap1GAP2 is a new GTPase-activating protein of Rap1 expressed in human platelets. Blood 2005; 105: 3185–3192.
Badner JA, Gershon ES . Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 2002; 7: 405–411.
Westin MA, Hunt MC, Alexson SE . Short- and medium-chain carnitine acyltransferases and acyl-CoA thioesterases in mouse provide complementary systems for transport of beta-oxidation products out of peroxisomes. Cell Mol Life Sci 2008; 65: 982–990.
Nigg JT . Temperament and developmental psychopathology. J Child Psychol Psychiatry 2006; 47: 395–422.
Cloninger CR, Van Eerdewegh P, Goate A, Edenberg HJ, Blangero J, Hesselbrock V et al. Anxiety proneness linked to epistatic loci in genome scan of human personality traits. Am J Med Genet 1998; 81: 313–317.
Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S et al. Genome-wide association scan of trait depression. Biol Psychiatry 2010; 68: 811–817.
de Moor MHM, Costa PT, Terracciano A, Krueger RF, de Geus EJC, Toshiko T et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry; e-pub ahead of print 21 December 2010; doi:10.1038/mp.2010.128 (in press).
Acknowledgements
We thank Ville Aalto and Irina Lisinen for their valuable help in data management. This study was supported by the Academy of Finland (Grant No. 123621 (LP-R)), the University of Helsinki Research Funds (Project No. 2106012 (LP-R)), the Turku University Hospital research grants (OTR), Tampere University Hospital Medical Fund (TL), Emil Aaltonen Foundation (TL), the Signe and Ane Gyllenberg Foundation (LK-J), the Alfred Kordelin Foundation (PM), the Otto A Malm Foundation (PM), the Jenny and Antti Wihuri Foundation (PM) and the National Doctoral Programme of Psychology in Finland (PM). MiKi is supported by the National Heart Lung and Blood Institute (R01HL36310), US, NIH: National Institute on Aging (R01AG013196; R01AG034454), US, NIH; the Academy of Finland, EU New OSH ERA research program, and the BUPA Foundation Specialist Research Grant, UK. The Young Finns study is also financially supported by the Academy of Finland (Grant No. 77841, 210283, 117832 and 121584), the Social Insurance Institution of Finland, Juho Vainio Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Disclaimer.The funding sources had neither been involved in the submission of the manuscript nor in the decision to publish the data.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Merjonen, P., Keltikangas-Järvinen, L., Jokela, M. et al. Hostility in adolescents and adults: a genome-wide association study of the Young Finns. Transl Psychiatry 1, e11 (2011). https://doi.org/10.1038/tp.2011.13
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2011.13
Keywords
This article is cited by
-
Conduct disorder
Nature Reviews Disease Primers (2019)
-
An integrated analysis of genes and functional pathways for aggression in human and rodent models
Molecular Psychiatry (2019)
-
High levels of cynical distrust partly predict premature mortality in middle-aged to ageing men
Journal of Behavioral Medicine (2017)
-
Serotonin receptor 1B genotype and hostility, anger and aggressive behavior through the lifespan: the Young Finns study
Journal of Behavioral Medicine (2013)
-
Meta-analysis of genome-wide association studies identifies common variants in CTNNA2 associated with excitement-seeking
Translational Psychiatry (2011)