Abstract
Staphylococcus sciuri is a bacterial pathogen associated with infections in animals and humans, and represents a reservoir for the mecA gene encoding methicillin-resistance in staphylococci. No S. sciuri siphophages were known. Here the identification and characterization of two temperate S. sciuri phages from the Siphoviridae family designated ϕ575 and ϕ879 are presented. The phages have icosahedral heads and flexible noncontractile tails that end with a tail spike. The genomes of the phages are 42,160 and 41,448 bp long and encode 58 and 55 ORFs, respectively, arranged in functional modules. Their head-tail morphogenesis modules are similar to those of Staphylococcus aureus ϕ13-like serogroup F phages, suggesting their common evolutionary origin. The genome of phage ϕ575 harbours genes for staphylokinase and phospholipase that might enhance the virulence of the bacterial hosts. In addition both of the phages package a homologue of the mecA gene, which is a requirement for its lateral transfer. Phage ϕ879 transduces tetracycline and aminoglycoside pSTS7-like resistance plasmids from its host to other S. sciuri strains and to S. aureus. Furthermore, both of the phages efficiently adsorb to numerous staphylococcal species, indicating that they may contribute to interspecies horizontal gene transfer.
Similar content being viewed by others
Introduction
Coagulase-negative and novobiocin-resistant Staphylococcus sciuri is mainly considered to be a commensal, animal-associated species with a broad range of habitats including domestic and wild animals, humans and the environment1,2. The presence of the bacteria correlates with animal diseases including dog dermatitis3, exudative epidermitis in pigs4, and with a number of nosocomial diseases of humans, namely endocarditis, pelvic inflammation, and wound infections5,6,7,8. It was speculated that the S. sciuri species group is a potential reservoir of virulence and antimicrobial resistance genes for other staphylococci9. Homologues of the mecA gene coding for a PBP2a-like protein responsible for methicillin resistance carried on the staphylococcal cassette chromosome mec (SCCmec) are often found in bacteria from the S. sciuri group10,11,12. The protein PBP2a of S. sciuri has 88% amino acid sequence identity to the PBP2a of methicillin-resistant S. aureus13. The acquisition of the SCCmec element by methicillin-susceptible S. aureus was crucial for the bacterium to become a successful pathogen14. It is hypothesized that S. aureus acquired this element directly from coagulase-negative staphylococci by transduction15. Transduction by temperate bacteriophages is a major path for the horizontal gene transfer of virulence and resistance determinants in staphylococci16,17,18,19,20. In recent years, the siphovirus genomic sequences of coagulase-negative staphylococci, which are important nosocomial pathogens, have been described in detail21,22,23,24.
Here the characterization of two newly discovered S. sciuri bacteriophages is presented and their potential for horizontal gene transfer of the SCC elements and antibiotic resistance plasmids is analysed.
Results
Identification of temperate phages in S. sciuri species and their host range
A screening method based on the UV-induction of prophages on a 96-well tissue culture plate was applied for rapid phage detection. From the set of 15 S. sciuri strains, only three temperate phages from strains P575, P879 and P581 were induced and were able to propagate on the suitable host strains P612, P723 and P583, respectively (Supplementary Table S1). Phages ϕ575 and ϕ879 lysed three and five strains, respectively, out of the 36 S. sciuri group strains tested (Supplementary Table S2). The third phage ϕ581 induced from S. sciuri P581, as well as phage ϕ879, also lysed a Staphylococcus lentus strain, thus demonstrating an interspecies host range in oxidase-positive and novobiocin-resistant staphylococci. However, a high-titre lysate of ϕ581 could not be obtained for further analyses.
Phage morphology and biological features
Both phages were able to propagate using the double-layer agar method, but not in liquid broth. They produced predominantly turbid plaques surrounded by a clear ring that indicates a high frequency of lysogenisation (Supplementary Fig. S1). Plaque diameter was variable from 0.8 to 1.4 mm. The indistinct edges of plaques that were similar for both phages may have been produced by the diffusion of lytic enzymes. The presence of endolysins in the phage lysate was confirmed by 1D SDS PAGE and subsequent mass spectrometry analysis (Fig. 1).
Left: silver-stained 1D SDS-PAGE gel with proteins extracted from purified phage particles. Selected bands were excised from the gel, digested with trypsin and subsequently analysed by mass spectrometry. Right: proteins identified by mass spectrometry with their theoretical molecular weight. M - molecular weight marker.
Morphological analysis by transmission electron microscopy (TEM) with negative staining and cryo-conditions (cryo-EM) revealed that both S. sciuri bacteriophages ϕ575 and ϕ879 belong to the Siphoviridae family. The phages consist of an icosahedral head (B1 morphology) with flexible, non-contractile tails ending with a tail spike (Fig. 2A and B). The diameters of the phage heads are 66 and 70 nm for ϕ575 and ϕ879, respectively (Supplementary Table S3). The electron density at the end of the tail spike suggests the presence of a metal ion (Fig. 2C).
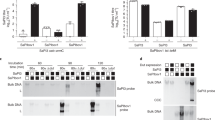
The adsorption kinetics of the phages were determined on various staphylococcal species including several S. aureus derivatives of laboratory isolates25,26,27,28 (Supplementary Table S1). Although both phages efficiently adsorbed onto other staphylococcal species (Fig. 3), they were unable to propagate on non-S. sciuri strains. The phages also adsorbed onto S. aureus RN4220 ΔtagO, which S. aureus siphoviruses are unable to adsorb onto due to the absence of wall teichoic acid (WTA). Phage ϕ575 adsorbed reversibly onto S. aureus PS 187. Both phages did not adsorb onto S. epidermidis CCM 2124 nor onto the S. aureus SA113 Δoat::Km mutant, which lacks the O-acetylation of the C6-OH of the muramic acid of peptidoglycan.
Genome structure
The phage genome sequences aligned with 100% identity to the prophage sequences of their lysogenic hosts. The genomes of ϕ575 and ϕ879 are arranged in functional modules and consist of 42,160 bp and 41,448 bp of dsDNA with a GC content of 31.8 and 32.1%, which is similar to the GC content of the host strains (32.4 and 32.3%). Both phage genomes have cohesive ends resulting in the formation of concatemers detected by pulsed-field gel electrophoresis (Supplementary Fig. S2). The physical ends of the phage DNA were not determined, but a significant decrease in DNA flexibility suggests that the physical ends of phage DNA are located in the region close to the 5′ end of the terminase small subunit (terS) gene.
Two additional prophages were identified in the strain S. sciuri P879. Their genomes are 43.3 and 51.0 kb in size and, like the characterized phages, have tyrosine integrase with a predicted XerC domain. Their genes encoding head proteins are arranged in a similar way to those in Mu-like phages29, however, attempts to induce these phages were unsuccessful.
Fifty-eight ORFs ranging from 156 bp to 4,971 bp were found in the ϕ575 genome, of which 37 have homologues in GenBank and the function was predicted for 33 of them (Supplementary Table S4). The gene density in this phage genome is 1.38 genes per kbp. The phage att site sequence ‘ATTCATCATAAAGTCAATACAGAACATTTGTACTTGTGCACAA’ was identified in the host strain P575 as a direct repeat between the ABC transporter uup gene (GenBank accession no. OFV61085.1) and a hypothetical protein gene (OFV61133.1). In the ϕ575 genome, downstream of the lytic module, the presence of virulence genes for staphylokinase (sak) and phospholipase (pla2) was detected.
The ϕ879 genome has 55 predicted ORFs ranging from 162 bp to 4,998 bp, of which 36 have homologues in GenBank and the function was predicted for 32 of them (Supplementary Table S4). Its gene density is 1.33 genes per kbp. The sequence of the phage att site is ‘GAATCCCTCCCAGGACGTAAATTACCAATATCCCGTTGTATCT’ and it is located between the genes for a hypothetical protein and the acetyltransferase from the GNAT family (OFV64881.1 and OFV64929.1), and overlaps with the tRNAArg gene by 17 nt (BFX02_03905). The integration module of both S. sciuri phages starts with the site-specific tyrosine recombinase XerC with an AP2-like DNA-binding integrase domain. The integrases themselves are dissimilar, with just 24% amino acid sequence identity.
Both phages have a σ-subunit of the RNA polymerase gene in their transcription regulation module. The presence of the sigma factor implies that both phages use their own promoters. The last protein in the DNA metabolism module is a 5-methylcytosine-specific restriction McrA-like protein that contains an HNHc endonuclease domain and is expected to provide higher resistance of their bacterial host to infection by other phages30. The presence of the HNH gene near the genes for the terminase subunits and portal protein is conserved in HK97-like phages31. The gene cluster responsible for replication exhibited similarities (66–73% nucleotide identity) to the genomes of S. aureus serogroup F phages such as ϕPVL or ϕN315. No predicted genes coding for functional RNAs were found in the ϕ575 and ϕ879 genomes.
The DNA packaging module follows the DNA metabolism module, and the two phages share 97% nucleotide identity in this module. The highest similarity (65–73% and 61–74% nucleotide identity) in the packaging and head module of the ϕ575 and ϕ879 phages was found to the S. aureus triple-converting phage ϕ13 from serological group F32 from the Sa3 integrase family, and the S. aureus Panton-Valentine converting phage ϕPVL33 from the Sa2 integrase family (Fig. 4). The module for head-tail morphogenesis encodes structural virion proteins and is also highly similar in both of the described phages (90% nucleotide identity).
The GenBank accession number for each sequence is given in brackets. Genomes were aligned using the blastn algorithm and similar regions with more than 65% identity are indicated. The positions and orientations of the coding regions are represented by arrows. Genome modules are color-coded according to the legend.
Structural proteins were analysed by SDS-PAGE in combination with mass spectrometry (Fig. 1). The major capsid protein (Mcp) belongs to the HK97 family. Its 120-amino acid-long N-terminal δ domain is cleaved by prohead protease and is not present in mature virions of the analysed phages. Mass spectrometry showed a 94% coverage of mature Mcp (Supplementary Fig. S3). The molecular weight of Mcp determined from SDS-PAGE gel is ~35 kDa (Fig. 1). Tail tape measure protein (Tmp), which determines the length of the tail, tends to be proteolytically cleaved34. The C-terminal end of Tmp contains a lysozyme-like domain involved in the hydrolysis of beta-1,4-linked polysaccharides of the bacterial cell wall and was detected in mature phages (Fig. 1). The function of the second largest protein in this module was predicted to be a tail spike protein determined on the basis of its position, length and secondary structure forming numerous beta sheets. No other tail-associated cell wall hydrolases were detected in the phage proteomes.
The module responsible for lysis of the bacterial cell is almost identical for both phages (Fig. 4). The holins of ϕ575 and ϕ879 are conserved in both phages and share over 90% amino acid identity. They exhibit more than 60% amino acid identity to S. aureus and Staphylococcus pseudintermedius prophage holins. The endolysins of ϕ575 and ϕ879 exhibit a low similarity to well-characterized staphylococcal endolysins and lack a cell-wall binding domain.
Horizontal transfer of pSTS7-like plasmid between S. sciuri and S. aureus strains
While analysing the sequencing data from purified phages, reads that map to bacterial and plasmid DNA sequences were found. These findings implied that phages are able to package non-phage DNA. Phage ϕ879 was able to package a 5,667-bp-long pSTS7-like plasmid designated pSSC723 from propagation strain P723. This plasmid encodes tetracycline efflux protein (TetL) and aminoglycoside adenyltransferase (AadD). The ratio between the plasmid-borne aadD gene and the reference tail protein gene determined by qPCR was 5.7 × 10−3. Plasmid pSSC723 belongs to the pC194 family and exhibits almost 100% nucleotide identity to the intergene spacer from the previously described S. epidermidis plasmid pSTS7. Plasmid pSTS7 originated from the interplasmid recombination of Bacillus subtilis plasmid pNS1981 and S. aureus plasmid pUB11035.
The transducing abilities of phage ϕ879 were verified by the transfer of pSSC723 to S. sciuri strains P600 and P574, and S. aureus strain RN4220. The donor strain P723 with plasmid pSSC723 grew on selective plates with tetracycline and kanamycin, whereas the recipients did not. Rare transductants with a frequency of about 10−11 were obtained in two intraspecies and one interspecies transduction to S. aureus. Plasmid identity in the donor strain and in transductants was confirmed by sequencing, and the genetic background of transductants was confirmed by macrorestriction analysis with SmaI by PFGE (Supplementary Fig. S4).
Packaging of S. sciuri SCCmec element
Both sequencing reads and quantification using real-time PCR proved the ability of phage ϕ879 to package mecA and plasmid genes from its bacterial host into virions. The packaging frequency of the mecA part of SCCmec was 3 × 10−5 as determined by qPCR analysis. Cassette chromosome recombinase (ccr) genes A, B and C were detected in phage ϕ879 particles using endpoint PCR. However, the successful transduction of SCCmec into recipient S. aureus strains was not observed.
A novel SCCmecP879 element whose parts packaged to phage ϕ879 virions was identified in methicillin-resistant prophage host S. sciuri strain P879. The SCCmecP879 element is 29,546 bp long, harbouring a mec A-class complex and ccrB3 and ccrA5 recombinases, and was integrated into the consensus insertion sequence site for SCC elements (GAAGCTTATCATAAGTAA). The major part (21 kb) of the SCCmecP879 shared 98% nucleotide identity with the SCCmec element of S. pseudintermedius strain KM24136 and the SCCmec element of Staphylococcus cohnii strain WC2837. However their J1 and J3 regions were different (Fig. 5). Ccr recombinases in SCCmecP879 exhibited 90–96% amino acid identity to recombinase complexes identified previously in S. sciuri38 or S. cohnii37. A cluster of genes including the cch gene for MCM-like helicase, which is conserved in a number of other SCC elements, was identified downstream of the recombinase genes ccrB3 and ccrA5. In addition, a non-mec SCC element designated SCCP879, which is 21,844 bp long, was identified adjacent to SCCmecP879. SCCP879 carries the ccrC recombinase gene, genes for a restriction-modification type I system and heavy metal resistance genes (Fig. 5).
Sequences of SCC elements from Staphylococcus pseudintermedius KM241, Staphylococcus sciuri P879 and Staphylococcus cohnii WC28 were aligned using the blastn algorithm and similar regions with more than 70% identity are indicated. The GenBank accession number for each sequence is given in brackets. The positions and orientations of coding regions are represented by arrows.
In methicillin-susceptible S. sciuri P575, a non-SCC homologue of the mecA gene (WP_070369365.1) was identified, coding for a PBP2a-like protein with 95% amino acid identity to PBP2a from S. aureus. This gene is located between a hypothetical protein gene (WP_058611716.1) and an amino acid permease gene (WP_070369378.1) and was also quantified in phage particles. The relative ratio between mecA and the reference tail protein gene copy numbers in phage particles was 8.2 × 10−4, which is higher than for phage ϕ879.
Discussion
Many prophages reside in the bacterial genomes and contribute to bacterial evolutionary processes via the horizontal exchange of genetic information40. Some prophages confer novel biological properties to their host strains, enabling them to adapt to new environments or obtain virulence determinants by lysogenic conversion, thereby driving bacterial adaptation and evolution. Typically, 1–4 prophages are found in each S. aureus genome41. Similarly, putative prophages often reside in the genomes of some coagulase-negative staphylococci42 and Macrococcus caseolyticus related to S. sciuri43, but only a few S. epidermidis, Staphylococcus hominis and Staphylococcus capitis viable phages have been genetically characterized in detail21,22,23. Recently, S. sciuri myoviruses isolated from urban sewage were described44, however no S. sciuri siphoviruses have been morphologically and genetically characterized to date.
The genomes of S. sciuri phages ϕ575 and ϕ879 have modular structures and sizes that are similar to those of other staphylococcal siphoviruses21,32,45,46. The phage genomes share 76% overall nucleotide identity, as calculated by a global alignment algorithm. Since they are related to S. aureus serogroup F phages belonging to integrase type Sa3, it is suggested that both bacteriophages belong to the Biseptimavirus genus47. Additional similarities with putative prophages across the genus Staphylococcus were identified in a Blast search of the microbial genome database. ϕ575 and ϕ879 exhibit 66–72% and 73–80% nucleotide identity, respectively, in their head-tail modules with prophages present in the genomes of S. pseudintermedius strains 063228 and NA4548 and Staphylococcus schleiferi strains 5909–02 and 2317–0349.
Even though the phages are similar, each integrates to different att sites in the host chromosome. This is because of differences in their integration modules. The integrase from ϕ879 has 51% amino acid identity to the Sa3 integrase from related S. aureus phage ϕ13, mainly in the C-terminal catalytic domain. The phage integrase from ϕ575 has 34% amino acid identity to the integrases from the Lysinibacillus and Bacillus spp. phages50.
Virulence factor-converting phages have not been detected in coagulase-negative staphylococci yet. Staphylokinase is a plasminogen activator protein secreted by most human lysogenic S. aureus strains. A secondary function of staphylokinase is its ability to neutralize host antimicrobial peptides whose binding may control its plasminogen activation properties52. The staphylokinase of ϕ575 exhibits a 52% amino acid sequence identity to that of S. aureus phages encoded by the immune evasion cluster associated with Sa3 integrase family phages53,54.
Phospholipase homologues can influence the innate and adaptive immune response and damage the membranes of host cells55. Phages carrying a gene for phospholipase were identified in streptococci56. Both virulence factors Sak and Pla2 contain an approximately 30-amino acid-long signal peptide that is required for protein secretion. The lysogenic hosts carrying phospholipase or staphylokinase are more likely to become virulent. There are two ORFs with opposite orientation between the sak and pla2 genes in the ϕ575 phage genome. The first, zipA-like gene was located on the complementary strand and the second gene has unknown function but was predicted to contain a transmembrane domain. The organisation of virulence factors in this region is similar to that of the immune evasion cluster, which harbours the sea, sak, scn, and chp genes54. Therefore the cluster of genes following the lysis module in ϕ575 could include other potential virulence factors.
Bacteriophages with a tail spike structure bind to proteinaceous receptors57. Prototypical phages with a tail spike are lactococcal bacteriophage p258 and B. subtilis phage SPP159. The high electron density observed at the end of the tail spike of ϕ575 and ϕ879 might be due to the presence of metal ions as was previously shown for the tail of bacteriophage p2, where iron, calcium and chloride ions were identified in the spike’s trimeric metal-binding structure60,61.
It is generally assumed that pac phages, which include the S. aureus phages of serogroup B, are responsible for generalized transduction62. In contrast, here it was shown that cos phages ϕ575 and ϕ879 package host genes in their virions. This finding corroborates previous observations of the transmission of pathogenicity islands via cos phages63,64.
Two types of mobile genetic elements, SCCmec and an pSTS7-like plasmid, were detected by qPCR in the phage ϕ879 particles. The SCCmecP879 core genes detected in ϕ879 virions are highly similar to SCC elements from other staphylococcal species such as S. pseudintermedius and S. cohnii. This indicates a possible horizontal interspecies transfer of this element. The MCM-like helicase identified in SCCmecP879 possibly participates in the extrachromosomal replication of this element65. This may increase the probability of SCCmec packaging into the phage virions. The relative quantity of packaged plasmid DNA in phage ϕ879 particles was higher than the relative quantity of the mecA gene from the SCCmec region. This implies that the pSSC723 plasmid is packaged as a multimer, as was suggested for other small plasmids66. In addition, plasmid interspecies and intraspecies transfer was proved at low frequencies, although the effective infection of S. sciuri phage ϕ879 has not been observed in the recipient strains. two recent studies, it was confirmed that phage adsorption on the recipient cell wall is sufficient for successful transduction, and effective infection is not necessary18,20.
The effective adsorption of analysed phages on the cells of non-S. sciuri species (Fig. 3) supports the speculated interspecies horizontal transfer of mobile elements or their parts by phages. A reversible adsorption of both phages was observed in S. aureus PS 187 that has a distinct structure of WTA28, suggesting the possibility of a two-stage adsorption. This type of adsorption has been described for the B. subtilis SPP1 phage as a result of a missing YueB membrane receptor on mutant host cells, where the glucosylated poly (glycerol-phosphate) of cell WTA proved to be a major target for SPP1 reversible binding67. A homologue of the YueB receptor described in B. subtilis was not found in staphylococci, however membrane proteins homologous to the phage infection proteins of the PIP family (e.g. YhgE) may act as possible receptors. The successful adsorption of ϕ575 and ϕ879 to S. aureus knockout mutant RN4220 ΔtagO and also the inability of both phages to adsorb onto S. aureus SA113 Δoat suggests that teichoic acids are not their only receptors, as with S. aureus phages68, but that O-acetyl groups at the 6-position of muramic acid play a role in the process of adsorption.
Conclusions
The identification of bacterial virulence factors encoded by S. sciuri phages, their ability to package and transmit mobile elements and to adsorb onto the cells of other staphylococcal species show that S. sciuri siphoviruses may contribute to the horizontal gene transfer within the Staphylococcus genus. Additionally, phages ϕ575 and ϕ879 exhibit evolutionary relationships to phages of the coagulase-positive species S. aureus and S. pseudintermedius, indicating a possible common gene pool of the phage genomes of these pathogens.
Material and Methods
Bacterial strains and Bacteriophages
Fifteen S. sciuri strains were selected for the screening of temperate phages and an additional 21 strains from the S. sciuri species group were used for the host-range determination. All S. sciuri strains were obtained from the National Reference Laboratory for Staphylococci (NRL/St), National Institute of Public Health (Prague, Czech Republic) and characterized previously2. The presence of the phages and their host range were tested on the set of S. sciuri strains by soft agar spot assay. The phage host strains, propagation strains, and strains used in the characterization of phage biological properties are listed in Supplementary Tables S1 and S2. The adsorption efficiencies of the phages were determined as described previously18. The antibiotic resistance pattern of selected S. sciuri strains was tested by the disc diffusion method on Mueller-Hinton agar (Oxoid) with discs (Oxoid) generally used for Gram-positive cocci (Supplementary Table S5).
High-throughput detection of UV-inducible prophages
A modified protocol designed for prophage induction on 96-well plates was used in this study69. Twenty μl of the overnight cultures were dispensed in a 96-well culture plate with 180 μl of CM1 nutrient broth (Oxoid, United Kingdom). The plates were incubated for 2 hours at 37 °C with shaking (120 rpm). After centrifugation at 1,100 g for 15 min, the pellet was resuspended in 200 μl of physiological solution and the plates were UV-irradiated for 25 s or 35 s using a 15 W UV-lamp (340 nm) at a distance of 60 cm. Forty μl of the irradiated suspension was transferred to a new plate together with 120 μl of physiological solution and 40 μl of 10× concentrated prophage broth per well, consisting of 10 g Trypton L42 (Oxoid), 2 g Yeast extract powder L21 (Oxoid), 2 g NaCl and 13 g Nutrient broth CM1 (Oxoid) dissolved in 100 ml of distilled water. The plates were protected from daylight and incubated at 37 °C for 2 hours. The plates were then centrifuged at 2,000 g for 10 min and the supernatants were passed through a 0.45 μm filter (TPP Techno Plastic Products, Switzerland) into a sterile microtube. If the screening resulted in lysis of the indicator strains, the presence of viable phages was proved by large-scale UV or mitomycin C induction.
Phage propagation and purification
Phages obtained from a single plaque were propagated using a double-layer agar technique with 1.5% 2YT bottom agar and 0.7% top Agar No. 1 L11 (Oxoid) with the addition of CaCl2 to a concentration of 2 mM. After overnight incubation, the top layer was disrupted and the phage was washed down with broth, centrifuged twice for 30 min at 3,100 g and filtered through a 0.45-μm filter. Phage particles were purified in a CsCl density gradient70.
Electron and Cryo-electron microscopy
Negative-stained samples were prepared by double staining in 2% uranyl acetate. Cryo samples were prepared by vitrification of the bacteriophage solution (at a concentration of 1010 PFU ml−1) on Quantifoil grids by plunging into liquid ethane using an FEI Vitrobot Mark IV. All samples were observed with an FEI Tecnai F20 electron microscope operated at 200 kV at a magnification of 29,000×.
Plasmid transduction
Intraspecies and interspecies transduction experiments were performed as described previously18. Transductants were selected on plates with kanamycin and tetracycline (both at concentration 8 μg ml−1), where the recipient strains S. sciuri P600, P574 and S. aureus RN4220 were unable to grow.
DNA extraction from the phage particles
DNase I (Sigma-Aldrich, Germany) and RNase A (Sigma-Aldrich) treatment was performed to remove any exogenous host genomic DNA and RNA from purified phage particles as described previously70. The DNA was isolated with a Phage DNA Isolation Kit (Norgen Biotek Corporation, Canada) according to the manufacturer’s recommendations. DNA from the viral particles for sequencing was isolated by phenol-chloroform extraction62. The concentration and purity of the phage DNA was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA).
Pulsed-field gel electrophoresis
One μg of phage DNA was loaded onto 1.5% agarose gel and separated by PFGE (Cheff Mapper, Bio-Rad, USA) to detect concatemerized cohesive ends. A constant voltage of 5 V cm−1 and switch times of 2-20 s with linear ramping were applied. Lambda DNA concatemers and a 5 kb DNA ladder (Bio-Rad) were used as molecular weight markers.
Genome sequencing and bioinformatic analysis
Phage genome and bacterial whole-genome shotgun (WGS) sequencing was performed using an Ion Torrent™ Personal Genome Machine (Ion PGM™). The purified genomic DNA was used for preparing a 400-bp sequencing library with an Ion Plus Fragment Library Kit (Thermo Fisher Scientific). The sample was loaded on a 316v2 chip and sequenced using an Ion PGM Hi-Q sequencing kit (Thermo Fisher Scientific). Quality trimming and error correction of the reads were performed with the Ion Torrent Suite Software (version 5.0.2). The assembly computation was performed using the implemented Assembler SPAdes (v.3.1.0) with default parameters for Ion Torrent data.
Sequences were manipulated and inspected in the cross-platform bioinformatics software Ugene v.1.23.171. The primal analysis of sequences was a combination of open reading frames (ORFs) prediction using GeneMark.hmm72 and automatic annotation by RAST73. Gene content was further examined via a BLASTp search on protein sequence databases74, CD-Search75 and InterPro v.5976. tRNAscan-SE77, RNAmmer v.1.278, and hmmsearch v.3.079 were used to analyse functional RNAs. Protein secondary structure was predicted with JPred480. Multiple sequence alignments were visualized using EasyFig v.2.181.
Nucleotide sequence accession numbers
The complete genomes of the S. sciuri phages ϕ575 and ϕ879 were deposited in GenBank under the accession numbers KY389063 and KY389064, respectively. The data from the WGS of bacterial host strains P575 and P879 were recorded in the GenBank WGS project under the accession numbers MDVU01 and MDVV01, respectively. The sequence of the pSSC723 plasmid was deposited in GenBank under the accession number KY389065.
SDS-PAGE and Mass Spectrometry
Vertical one-dimensional electrophoresis (1-DE), using Bio-Rad equipment, was performed in a Protean II xi Cell (discontinuous 12% T SDS-PAGE). Precision Plus Protein Standard (Bio-Rad) was applied as the molecular weight marker. Proteins were stained with a ProteoSilver Plus kit (Sigma Aldrich). Protein bands were excised, destained and subjected to tryptic digestion (40 °C, 2 h) without the reduction and alkylation of cysteine residues. Digested peptides were extracted from gels using 50% acetonitrile solution with 2.5% formic acid and concentrated in a SpeedVac concentrator (Thermo Fisher Scientific). LC-MS/MS analyses of the peptide mixture were done using an RSLCnano system (Thermo Fisher Scientific) on-line connected to an Impact II Ultra-High Resolution Qq-Time-Of-Flight mass spectrometer (Bruker, Bremen, Germany). Prior to LC separation, tryptic digests were on-line concentrated in a trap column. The peptides were separated using an Acclaim Pepmap100 C18 column (3 μm particles, 75 μm × 500 mm; Thermo Fisher Scientific, 300 nl min−1) and a 0.1% FA/acetonitrile gradient. MS data were acquired in a data-dependent strategy with a 3 s cycle time. Mass range was set to 150–2,200 m/z and precursors were selected from 300 to 2,000 m/z. Mascot (version 2.4.1) MS/MS ion searches were done against a local database containing translated phage insert sequences. Mass tolerance for precursors and MS/MS fragments were 15 ppm and 0.05 Da, respectively. The oxidation of methionine, deamidation (N, Q) and propionamide (C) were set as variable modifications for all searches.
The relative quantification analysis of bacterial genes in phage particles
The target genes for qPCR were first detected by conventional end-point PCR and the amplicons verified by sequencing. A LightCycler 490 Real-Time PCR Instrument (Roche Diagnostics, USA) was used for relative quantification analysis. Reactions were carried out in triplicates in MicroAmp® optical 96-well reaction plates sealed with optical adhesive covers (Roche Diagnostics). Each reaction mixture (10 μl) contained 7.5 μl FastStart® TaqMan Probe Master (Roche Diagnostics), 900 nM of each primer, 250 nM of hydrolysis probe (Supplementary Table S6), and 50 ng of template DNA. An initial denaturation of DNA at 95 °C for 10 min was followed by 40 cycles of amplification (95 °C for 15 s and 60 °C for 45 s).
The relative quantification analysis was performed as a dual-colour experiment. The FAM probes were used to target mecA and aminoglycoside acetyltransferase genes, the Cy5 probe was used for reference phage gene coding for the head-tail connector protein. The results were generated by the Light-Cycler software using the maximum of the second derivative, and a target gene was paired with the reference by a one-to-one pairing analysis. The reaction efficiency was calculated for each analysed gene, and the results were normalized according to the efficiencies from the relative quantification. The calculated ratio between the number of target gene copies and reference gene copies was estimated.
Additional Information
How to cite this article: Zeman, M. et al. Staphylococcus sciuri bacteriophages double-convert for staphylokinase and phospholipase, mediate interspecies plasmid transduction, and package mecA gene. Sci. Rep. 7, 46319; doi: 10.1038/srep46319 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Kloos, W. E., Schleifer, K. H. & Smith, R. F. Characterization of Staphylococcus sciuri sp. nov. and its subspecies. International Journal of Systematic Bacteriology 26, 22–37, doi: 10.1099/00207713-26-1-22 (1976).
Švec, P., Petráš, P., Pantůček, R., Doškař, J. & Sedláček, I. High intraspecies heterogeneity within Staphylococcus sciuri and rejection of its classification into S. sciuri subsp. sciuri, S. sciuri subsp. carnaticus and S. sciuri subsp. rodentium . International Journal of Systematic and Evolutionary Microbiology 66, 5181–5186, doi: 10.1099/ijsem.0.001493 (2016).
Hauschild, T. & Wojcik, A. Species distribution and properties of staphylococci from canine dermatitis. Research in Veterinary Science 82, 1–6, doi: 10.1016/j.rvsc.2006.04.004 (2007).
Chen, S. et al. A highly pathogenic strain of Staphylococcus sciuri caused fatal exudative epidermitis in piglets. PLoS One 2, e147, doi: 10.1371/journal.pone.0000147 (2007).
Hedin, G. & Widerstrom, M. Endocarditis due to Staphylococcus sciuri . European Journal of Clinical Microbiology and Infectious Diseases 17, 673–675, doi: 10.1007/BF01708356 (1998).
Stepanović, S., Dakić, I., Djukić, S., Lozuk, B. & Svabic-Vlahović, M. Surgical wound infection associated with Staphylococcus sciuri . Scandinavian Journal of Infectious Diseases 34, 685–686, doi: 10.1080/00365540110076949a (2002).
Dakić, I. et al. Isolation and molecular characterization of Staphylococcus sciuri in the hospital environment. Journal of Clinical Microbiology 43, 2782–2785, doi: 10.1128/JCM.43.6.2782-2785.2005 (2005).
Stepanović, S., Ježek, P., Dakić, I., Vuković, D. & Seifert, L. Staphylococcus sciuri: an unusual cause of pelvic inflammatory disease. International Journal of STD & AIDS 16, 452–453, doi: 10.1258/0956462054093999 (2005).
Nemeghaire, S. et al. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Veterinary Microbiology 171, 342–356, doi: 10.1016/j.vetmic.2014.02.005 (2014).
Couto, I., Wu, S. W., Tomasz, A. & de Lencastre, H. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. Journal of Bacteriology 185, 645–653, doi: 10.1128/JB.185.2.645-653.2003 (2003).
Rolo, J., de Lencastre, H. & Miragaia, M. High frequency and diversity of cassette chromosome recombinases (ccr) in methicillin-susceptible Staphylococcus sciuri . Journal of Antimicrobial Chemotherapy 69, 1461–1469, doi: 10.1093/jac/dku028 (2014).
Zhou, Y., Antignac, A., Wu, S. W. & Tomasz, A. Penicillin-binding proteins and cell wall composition in beta-lactam-sensitive and -resistant strains of Staphylococcus sciuri . Journal of Bacteriology 190, 508–514, doi: 10.1128/JB.01549-07 (2008).
Wu, S., de Lencastre, H. & Tomasz, A. Genetic organization of the mecA region in methicillin-susceptible and methicillin-resistant strains of Staphylococcus sciuri . Journal of Bacteriology 180, 236–242, doi: 10.1128/JB.180.2.236-242.1998 (1998).
Robinson, D. A. & Enright, M. C. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus . Antimicrobial Agents and Chemotherapy 47, 3926–3934, doi: 10.1128/AAC.47.12.3926-3934.2003 (2003).
Otto, M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection. Bioessays 35, 4–11, doi: 10.1002/bies.201200112 (2013).
Varga, M. et al. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiology Letters 332, 146–152, doi: 10.1111/j.1574-6968.2012.02589.x (2012).
Scharn, C. R., Tenover, F. C. & Goering, R. V. Transduction of staphylococcal cassette chromosome mec elements between strains of Staphylococcus aureus . Antimicrobial Agents and Chemotherapy 57, 5233–5238, doi: 10.1128/AAC.01058-13 (2013).
Mašlaňová, I., Stříbná, S., Doškař, J. & Pantůček, R. Efficient plasmid transduction to Staphylococcus aureus strains insensitive to the lytic action of transducing phage. FEMS Microbiology Letters 363, fnw211, doi: 10.1093/femsle/fnw211 (2016).
Stanczak-Mrozek, K. I. et al. Within-host diversity of MRSA antimicrobial resistances. Journal of Antimicrobial Chemotherapy 70, 2191–2198, doi: 10.1093/jac/dkv119 (2015).
Haaber, J. et al. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nature Communications 7, 13333, doi: 10.1038/ncomms13333 (2016).
Deghorain, M. et al. Characterization of novel phages isolated in coagulase-negative staphylococci reveals evolutionary relationships with Staphylococcus aureus phages. Journal of Bacteriology 194, 5829–5839, doi: 10.1128/JB.01085-12 (2012).
Daniel, A., Bonnen, P. E. & Fischetti, V. A. First complete genome sequence of two Staphylococcus epidermidis bacteriophages. Journal of Bacteriology 189, 2086–2100, doi: 10.1128/JB.01637-06 (2007).
Gutiérrez, D., Martínez, B., Rodríguez, A. & García, P. Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential. BMC Genomics 13, 228, doi: 10.1186/1471-2164-13-228 (2012).
Melo, L. D. et al. Characterization of Staphylococcus epidermidis phage vB_SepS_SEP9 - a unique member of the Siphoviridae family. Research in Microbiology 165, 679–685, doi: 10.1016/j.resmic.2014.09.012 (2014).
Kreiswirth, B. N. et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712, doi: 10.1038/305709a0 (1983).
Xia, G. et al. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. Journal of Bacteriology 193, 4006–4009, doi: 10.1128/JB.01412-10 (2011).
Bera, A., Herbert, S., Jakob, A., Vollmer, W. & Götz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus . Molecular Microbiology 55, 778–787, doi: 10.1111/j.1365-2958.2004.04446.x (2005).
Winstel, V., Sanchez-Carballo, P., Holst, O., Xia, G. & Peschel, A. Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. mBio 5, e00869, doi: 10.1128/mBio.00869-14 (2014).
Morgan, G. J., Hatfull, G. F., Casjens, S. & Hendrix, R. W. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus . Journal of Molecular Biology 317, 337–359, doi: 10.1006/jmbi.2002.5437 (2002).
Campbell, A. M. In Bacterial Genomes: Physical Structure and Analysis(eds F. J. de Bruijn, J. R. Lupski & G. M. Weinstock ) Ch. Prophages and Cryptic Prophages 23–29 (Springer, 1998).
Moodley, S., Maxwell, K. L. & Kanelis, V. The protein gp74 from the bacteriophage HK97 functions as a HNH endonuclease. Protein Science 21, 809–818, doi: 10.1002/pro.2064 (2012).
Iandolo, J. J. et al. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene 289, 109–118, doi: 10.1016/S0378-1119(02)00481-X (2002).
Kaneko, J., Kimura, T., Kawakami, Y., Tomita, T. & Kamio, Y. Panton-valentine leukocidin genes in a phage-like particle isolated from mitomycin C-treated Staphylococcus aureus V8 (ATCC 49775). Bioscience Biotechnology and Biochemistry 61, 1960–1962, doi: 10.1271/bbb.61.1960 (1997).
Tsui, L. C. & Hendrix, R. W. Proteolytic processing of phage lambda tail protein gpH: timing of the cleavage. Virology 125, 257–264, doi: 10.1016/0042-6822(83)90199-X (1983).
Schwarz, S., Gregory, P. D., Werckenthin, C., Curnock, S. & Dyke, K. G. A novel plasmid from Staphylococcus epidermidis specifying resistance to kanamycin, neomycin and tetracycline. Journal of Medical Microbiology 45, 57–63, doi: 10.1099/00222615-45-1-57 (1996).
Descloux, S., Rossano, A. & Perreten, V. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius . Journal of Clinical Microbiology 46, 1818–1823, doi: 10.1128/JCM.02255-07 (2008).
Zong, Z. & Lu, X. Characterization of a new SCCmec element in Staphylococcus cohnii . PLoS One 5, e14016, doi: 10.1371/journal.pone.0014016 (2010).
Urushibara, N., Paul, S. K., Hossain, M. A., Kawaguchiya, M. & Kobayashi, N. Analysis of Staphylococcal cassette chromosome mec in Staphylococcus haemolyticus and Staphylococcus sciuri: identification of a novel ccr gene complex with a newly identified ccrA allotype (ccrA7). Microbial Drug Resistance 17, 291–297, doi: 10.1089/mdr.2010.0144 (2011).
Harrison, E. M. et al. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri . Journal of Antimicrobial Chemotherapy 69, 911–918, doi: 10.1093/jac/dkt452 (2014).
Casjens, S. Prophages and bacterial genomics: what have we learned so far? Molecular Microbiology 49, 277–300, doi: 10.1046/j.1365-2958.2003.03580.x (2003).
Goerke, C. et al. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. Journal of Bacteriology 191, 3462–3468, doi: 10.1128/JB.01804-08 (2009).
Takeuchi, F. et al. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. Journal of Bacteriology 187, 7292–7308, doi: 10.1128/JB.187.21.7292-7308.2005 (2005).
Baba, T. et al. Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. Journal of Bacteriology 191, 1180–1190, doi: 10.1128/JB.01058-08 (2009).
Jurczak-Kurek, A. et al. Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Scientific Reports 6, 34338, doi: 10.1038/srep34338 (2016).
Kwan, T., Liu, J., DuBow, M., Gros, P. & Pelletier, J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proceedings of the National Academy of Sciences of the USA 102, 5174–5179, doi: 10.1073/pnas.0501140102 (2005).
Kahánková, J. et al. Multilocus PCR typing strategy for differentiation of Staphylococcus aureus siphoviruses reflecting their modular genome structure. Environmental Microbiology 12, 2527–2538, doi: 10.1111/j.1462-2920.2010.02226.x (2010).
Gutiérrez, D. et al. Three proposed new bacteriophage genera of staphylococcal phages: “3Alikevirus”, “77likevirus” and “Phietalikevirus”. Archives of Virology 159, 389–398, doi: 10.1007/s00705-013-1833-1 (2014).
Riley, M. C., Perreten, V., Bemis, D. A. & Kania, S. A. Complete genome sequences of three important methicillin-resistant clinical isolates of Staphylococcus pseudintermedius . Genome Announcements 4, e01194-16, doi: 10.1128/genomeA.01194-16 (2016).
Misic, A. M., Cain, C. L., Morris, D. O., Rankin, S. C. & Beiting, D. P. Complete genome sequence and methylome of Staphylococcus schleiferi, an important cause of skin and ear infections in veterinary medicine. Genome Announcements 3, e01011-15, doi: 10.1128/genomeA.01011-15 (2015).
Xu, K., Yuan, Z., Rayner, S. & Hu, X. Genome comparison provides molecular insights into the phylogeny of the reassigned new genus Lysinibacillus. BMC Genomics 16, 140, doi: 10.1186/s12864-015-1359-x (2015).
Kurata, A., Nishimura, M., Kishimoto, N. & Kobayashi, T. Draft genome sequence of a deep-sea bacterium, Bacillus niacini strain JAM F8, involved in the degradation of glycosaminoglycans. Genome Announcements 2, e00983-14, doi: 10.1128/genomeA.00983-14 (2014).
Nguyen, L. T. & Vogel, H. J. Staphylokinase has distinct modes of interaction with antimicrobial peptides, modulating its plasminogen-activation properties. Scientific Reports 6, 31817, doi: 10.1038/srep31817 (2016).
Coleman, D. C. et al. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. Journal of General Microbiology 135, 1679–1697, doi: 10.1099/00221287-135-6-1679 (1989).
van Wamel, W. J., Rooijakkers, S. H., Ruyken, M., van Kessel, K. P. & van Strijp, J. A. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. Journal of Bacteriology 188, 1310–1315, doi: 10.1128/JB.188.4.1310-1315.2006 (2006).
Sitkiewicz, I., Stockbauer, K. E. & Musser, J. M. Secreted bacterial phospholipase A2 enzymes: better living through phospholipolysis. Trends in Microbiology 15, 63–69, doi: 10.1016/j.tim.2006.12.003 (2007).
Beres, S. B. et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proceedings of the National Academy of Sciences of the USA 99, 10078–10083, doi: 10.1073/pnas.152298499 (2002).
Mahony, J. & van Sinderen, D. Structural aspects of the interaction of dairy phages with their host bacteria. Viruses 4, 1410–1424, doi: 10.3390/v4091410 (2012).
Sciara, G. et al. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proceedings of the National Academy of Sciences of the USA 107, 6852–6857, doi: 10.1073/pnas.1000232107 (2010).
Vinga, I. et al. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Molecular Microbiology 83, 289–303, doi: 10.1111/j.1365-2958.2011.07931.x (2012).
Browning, C., Shneider, M. M., Bowman, V. D., Schwarzer, D. & Leiman, P. G. Phage pierces the host cell membrane with the iron-loaded spike. Structure 20, 326–339, doi: 10.1016/j.str.2011.12.009 (2012).
Yamashita, E. et al. The host-binding domain of the P2 phage tail spike reveals a trimeric iron-binding structure. Acta Crystallographica Sect. F Structural Biology and Crystallization Communications 67, 837–841, doi: 10.1107/S1744309111005999 (2011).
Doškař, J. et al. Genomic relatedness of Staphylococcus aureus phages of the International Typing Set and detection of serogroup A, B, and F prophages in lysogenic strains. Canadian Journal of Microbiology 46, 1066–1076, doi: 10.1139/w00-097 (2000).
Chen, J. et al. Intra- and inter-generic transfer of pathogenicity island-encoded virulence genes by cos phages. ISME Journal 9, 1260–1263, doi: 10.1038/ismej.2014.187 (2015).
Quiles-Puchalt, N. et al. Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases. Proceedings of the National Academy of Sciences of the USA 111, 6016–6021, doi: 10.1073/pnas.1320538111 (2014).
Mir-Sanchis, I. et al. Staphylococcal SCCmec elements encode an active MCM-like helicase and thus may be replicative. Nature Structural & Molecular Biology 23, 891–898, doi: 10.1038/nsmb.3286 (2016).
Novick, R. P., Edelman, I. & Lofdahl, S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. Journal of Molecular Biology 192, 209–220, doi: 10.1016/0022-2836(86)90360-8 (1986).
Baptista, C., Santos, M. A. & Sao-Jose, C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. Journal of Bacteriology 190, 4989–4996, doi: 10.1128/JB.00349-08 (2008).
Li, X. et al. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus . Scientific Reports 6, 26455, doi: 10.1038/srep26455 (2016).
McDonald, J. E., Smith, D. L., Fogg, P. C., McCarthy, A. J. & Allison, H. E. High-throughput method for rapid induction of prophages from lysogens and its application in the study of Shiga Toxin-encoding Escherichia coli strains. Applied and Environmental Microbiology 76, 2360–2365, doi: 10.1128/AEM.02923-09 (2010).
Mašlaňová, I. et al. Bacteriophages of Staphylococcus aureus efficiently package various bacterial genes and mobile genetic elements including SCCmec with different frequencies. Environmental Microbiology Reports 5, 66–73, doi: 10.1111/j.1758-2229.2012.00378.x (2013).
Okonechnikov, K., Golosova, O., Fursov, M. & team, U. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167, doi: 10.1093/bioinformatics/bts091 (2012).
Besemer, J. & Borodovsky, M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Research 33, W451–454, doi: 10.1093/nar/gki487 (2005).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75, doi: 10.1186/1471-2164-9-75 (2008).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Research 43, D222–226, doi: 10.1093/nar/gku1221 (2015).
Mitchell, A. et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Research 43, D213–221, doi: 10.1093/nar/gku1243 (2015).
Schattner, P., Brooks, A. N. & Lowe, T. M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Research 33, W686–689, doi: 10.1093/nar/gki366 (2005).
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research 35, 3100–3108, doi: 10.1093/nar/gkm160 (2007).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Research 39, W29–37, doi: 10.1093/nar/gkr367 (2011).
Drozdetskiy, A., Cole, C., Procter, J. & Barton, G. J. JPred4: a protein secondary structure prediction server. Nucleic Acids Research 43, W389–394, doi: 10.1093/nar/gkv332 (2015).
Sullivan, M. J., Petty, N. K. & Beatson, S. A. Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010, doi: 10.1093/bioinformatics/btr039 (2011).
Acknowledgements
This work was supported by a grant from the Czech Science Foundation (GP13-05069P). CIISB research infrastructure project LM2015043 funded by MEYS CR is gratefully acknowledged for financial support of the measurements at the Proteomics CF and the Cryo-electron Microscopy and Tomography CF. We wish to thank Hana Konečná for the SDS-PAGE separation of phage protein isolates.
Author information
Authors and Affiliations
Contributions
M.Z., I.M., A.I. and R.P. isolated the phages and designed the study. Horizontal gene transfer experiments were performed by I.M. and A.I.; M.Š. and P.P. performed the electron microscopy experiments. Whole-genome sequencing was performed by M.Z., K.B. and V.V.; A.I. and M.Z. analysed the data. K.M. and Z.Z. were responsible for proteomic analysis. M.Z., I.M., J.D., P.P. and R.P. wrote the manuscript. All the authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zeman, M., Mašlaňová, I., Indráková, A. et al. Staphylococcus sciuri bacteriophages double-convert for staphylokinase and phospholipase, mediate interspecies plasmid transduction, and package mecA gene. Sci Rep 7, 46319 (2017). https://doi.org/10.1038/srep46319
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep46319
This article is cited by
-
Nasotracheal Microbiota of Nestlings of Parent White storks with Different Foraging Habits in Spain
EcoHealth (2023)
-
Multi-species host range of staphylococcal phages isolated from wastewater
Nature Communications (2021)
-
Phage Transduction is Involved in the Intergeneric Spread of Antibiotic Resistance-Associated blaCTX-M, mel, and tetM Loci in Natural Populations of Some Human and Animal Bacterial Pathogens
Current Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.