Abstract
Huntingtin interacting protein 1 (HIP1) is an endocytic protein which is overexpressed in a variety of human cancers and involved in cancer-causing translocation in leukemia. However, the prognostic impact of HIP1 expression on AML remains unclear. In this study, quantification of HIP1 transcript by real-time quantitative PCR in bone marrow blasts was performed in 270 AML patients. As a result, high HIP1 expression was seen more frequently in older patients, M4/M5 morphology and genes of NPM1 and DNMT3A mutations, and underrepresented in favorable karyotype subgroups and CEBPA double allele mutations in our AML patients. We also found high HIP1 expressers showed lower levels of hemoglobin. In addition, overexpression of HIP1 was associated with an inferior overall survival. The prognostic value of HIP1 expression was validated in patients from an independent TCGA cohort. Notably, up-regulation of miR-16, miR-15a, miR-28 and miR-660 were seen in high HIP1 expressers from the two independent cohorts. In vitro, interfereing of HIP1 expression by siRNA suppressed the proliferation of leukemic cells, and downregulation of these miRNAs were seen in THP-1 and Kasumi cell lines after silencing HIP1 expression. In conclusion, the HIP1 gene expression might serve as a reliable predictor for overall survival in AML patients.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of hematologic malignancies with various genetic abnormalities and variable responses to treatment. To date, AML patients can be classified into three risk subgroups according to karyotype abnormalities: favorable, intermediate and adverse. In addition to chromosome lesions, several genes such as NPM1, FLT3-ITD and CEBPA mutations have been recommended as reliable prognostic factors1. However, only half of AML patients obtained cytogenetic abnormalities. Thus, reliable biomarkers are still required in clinical practice2.
With the advent of the high throughput transcriptomic profiling, biomarker identification has been taken to the genomic level3. Although multiple genes, particularly signaling pathways, provide a stronger and more reliable prognostic assessment, prognostic effects must first be studied at the individual gene level. This is because such an analysis will provide a rationale for mechanistic studies followed by therapeutic targeting. The hypothesis is that one disrupted gene was enough to regulate the relevant signaling pathway, leading to leukemia cell proliferation or metastasis. Thus, in order to identify such a driver gene, we searched for public articles about pathway analysis using gene microarray for AML patients. Fortunately, we found that huntington’s disease (HD) signaling pathway is one of the most significant pathways changed in AML blasts compared with normal CD34 bone marrow samples as previously reported4. We therefore focused on the HD signaling pathway to uncover the underlying oncogene. It is believed that mutant huntingtin (HTT) protein causes HD. One of the mechanisms is mutant HTT protein leads to decreased binding affinity for HTT interacting protein 1 (HIP1), thus causing disruption of HIP1’s normal function, and also accumulation of high levels of the free form of HIP15. HIP1 contains evolutionarily conserved sequences, including a leucine zipper motif and a carboxyl terminus with homology to TALIN, a cytoskeletal actin binding protein6. Although the true function of HIP1 remains unknown, it has been shown HIP1 protein has a role in the clathrin-mediated endocytosis which regulates several different signaling pathways, receptor trafficking and cytoskeleton dynamics7. Notably, it has previously reported alterations in HIP1 protein have been associated with tumors. Analysis by western blot showed more than 50 cancer cell lines had high levels of HIP1 protein7. Similarly, overexpression of HIP1 gene was also observed in multiple human cancers including prostate cancer8, breast cancer9, brain tumor10, Merkel cell carcinomas11 and lymphoma12. Furthermore, in vitro analysis of the effects of HIP1 overexpression on cells indicated that it can transform fibroblasts9. These results suggest HIP1 acts as an oncogene in solid tumors. With respect to hematopoietic malignancies, the first clue that HIP1 might have a role in tumorigenesis came in 1988, when the fusion of HIP1 and platelet-derived growth factor receptor was discovered as the cause of a chronic myelomonocytic leukemia13. Taken together, HIP1 expression might serve as a useful biomarker in AML owing to the oncogenic propensity. However, the biological feature and prognostic value of HIP1 expression in AML blasts remains unclear.
Here, we found AML patients with high HIP1 expression had a distinct microRNA signature and poor survival in our large cohort of patients. The prognostic value of HIP1 expression was also validated in an independent cohort of AML patients. This study provides a reliable prognostic biomarker and critical drug target for AML patients.
Results
Characteristics of patients with high HIP1 expression
The distribution of HIP1 expression was binormal and exhibited two clusters low and high expressers (Figure S1). The cutoff value was estimated using Cutoff Finder software analysis. Thus, 90 (33%) were classified as low and 180 (67%) high HIP1 expression. Clinical characteristics of patients with high HIP1 expression are summarized in Table 1. High expressers were older (P = 0.013), had lower hemoglobin levels (P = 0.026), and were more frequently in AML FAB subtype M4 (12% vs. 3%) and M5 (28% vs. 18%) morphology (P = 0.025) than low expressers. Patients with high HIP1 expression were associated with a significantly higher frequency of NPM1 mutations (30% vs. 17%, P = 0.036) and DNMT3A mutations (13% vs. 5%, P = 0.031), a significantly lower frequency of favorable karyotype risk subgroup (2% vs. 9%, P = 0.037), CEBPA double allele mutations (7% vs. 24%, P < 0.001), compared with patients with low expression. There was no statistically significant correlation between HIP1 expression and other variables including sex, white blood cell counts (WBC), platelet counts, percentage of bone marrow blasts and FLT3-ITD positive and treatment protocols (Table 1).
Association of HIP1 expression with clinical outcome from the ZIH cohort
With a median follow-up for living patients of 484 days with 95% confidence interval 374–1262 days, high HIP1 expressers (n = 180) had more adverse OS compared to low expressers (n = 90) (Fig. 1A). Importantly, in the subgroup analyses we found high HIP1 expressions were associated with poor OS in patients with both cytogenetic intermediate risk group and cytogenetically normal AML (Figure S2A,B). In order to identify the potential confounders or interactive factors, we conducted stratified analyses and interactive analyses. As shown in Supplementary Figure S3, there were no significant interactions among these factors. Even if we taken these factors as confounders, HIP1 expression was still as an independent prognostic factor in multivariate analysis after adjusting for age, WBC, karyotype risk groups, and genes of FLT3-ITD, NPM1, CEBPA and DNMT3A mutations [for OS HR (95%CI), 1.658 (1.068, 2.576); P = 0.024; Table 2]. Moreover, we also conducted landmark analysis by including patients whose survival was more than 30 days in order to ignore the cause of induction death by intense chemotherapy. As a result, high expression of HIP1 was still independently associated with poor OS [HR (95%CI), 1.766(1.074, 2.905), P = 0.025] in the multivariate survival analyses (Table S1).
With respect to the induction remission rate, high expressers had lower complete remission rate compared with low expressers in univariate analysis [OR (95%CI), 0.491(0.256,0.903), P = 0.026, Table S2]. However, the significance did not stand after adjustment with other factors like age, WBC, cytogenetic risk groups, genes of FLT3-ITD, NPM1, CEBPA and DNMT3A mutations and treatment protocols in the multivariate analysis (Table S2).
Validation of the impact of HIP1 expression on survival from the TCGA cohort
In order to validate the prognostic values of HIP1 expression in our cohort, we defined low HIP1 expressers from TCGA cohort using the same percentage (33%) of low expressers based on the same estimated method as mentioned in our cohort (Figure S1B). Correspondingly, of 197 patients from the TCGA cohort, 66 (33%) were defined as low HIP1 expressers and 131 (67%) as high expressers. Characteristics of patients with high HIP1 expression from the TCGA cohort were described in Supplementary Table S3. Similarly, high HIP1 expressers were predominant in older patients (P = 0.04), more frequently in M4 (29% vs. 5%) and M5 (15% vs. 5%) morphology (P < 0.001), had a significantly higher frequency of NPM1 mutations (36% vs.11%, P < 0.001) and DNMT3A mutations (32% vs. 12%, P = 0.003), and had a significantly lower frequency of favorable karyotype risk subgroup (11% vs. 33%, P < 0.001), compared with low expressers (Table S3). In contrast with our cohort, we also found high expressers had higher levels of WBC and a higher frequency of FLT3-ITD positive. There are no differences between high and low expressers with respect to sex, percent BM blasts, genes of CEBPA, IDH1, IDH2 mutations and consolidation therapy such as bone marrow transplantations.
To test whether HIP1 was a robust biomarker among gene expression profiles of AML in the TCGA cohort, we carried out the resampling statistics using the method of multiple survival screening (MSS)14. As a result, 45 genes whose q-values are less than 0.05 were identified as survival genes. As expected, HIP1 was observed as a robust biomarker in the top 30 most frequent genes among the predictive random gene sets (Table S4). In univariate analysis, patients with high HIP1 expressers were associated with adverse OS compared with lower expressers (Fig. 1B). Moreover, in multivariate analysis, high HIP1 expressers were significantly associated with poor OS [HR, 1.558(1.017, 2.385); P = 0.041, Table S5] in the context of age, WBC, karyotype-risk groups and genes mutations of FLT3-ITD, NPM1, CEBPA, DNMT3A, IDH1, IDH2, and bone marrow transplantation.
MicroRNA expression profiling
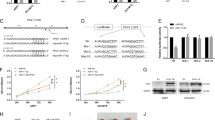
We applied the propensity score analysis to match each CN-AML patient with high and low HIP1 in our cohort, matching was based on age, WBC, cytogenetic group and genes of FLT3-ITD, NPM1, CEBPA, DNMT3A mutations which might affect the microRNA expression (Table S6). Thus, we selected six samples with high and five matching samples with low HIP1 expression to assess the differences of microRNA (miR) expression. The most significant changes of miRs in high expressers included up-regulation of miR-146b-5p, miR-16, miR-361-3p, miR-26a, miR-197, miR-28-5p, miR-590-5p, miR-140-5p, miR-185, miR-22, miR-17, miR-15a, miR-4306, let-7a, miR-130b, miR-660 and miR-338-3p and down-regulation of miR-4270, miR-3663-3p, hsv1-miR-H18, hsv2-miR-H6, miR-3665, miR-1225-5p, miR-1275, miR-1915, miR-3196, miR-3198, miR-3648 and miR-718 (p-value < 0.05, Fig. 2). Among these miRs, up-regulation of miR-16, miR-15a, miR-28 and miR-660 were also seen significantly changes in high HIP1 expressers in a large and independent cohort of TCGA patients (Fig. 3).
siRNA interference
The relevance of high levels of HIP1 expression to the poor survival suggests that HIP1 may be functionally important for maintaining the continuous proliferation of leukemia cells. To example this possibility, we measured the proliferation of Kasumi-1 and THP-1 cells using siRNA to silence HIP1 expression. Quantitative RT-PCR analysis showed that siRNA treatment resulted in approximately 83% and 63% knockdown of HIP1 mRNA expression in THP-1 and Kasumi-1 leukemia cells after 72 hours compared to negative control (Figure S4). It also led to a significant decreased growth of THP-1 and Kasumi-1 cells (Fig. 4). Importantly, HIP1 interference in THP-1 cell line dramatically reduced the expression of miR-16, miR-15a, miR-28 and miR-660 (Fig. 5A). In parallel, silencing HIP1 expression in Kasumi-1 cell line significantly reduced the expression of these miRNAs (Fig. 5B). These results suggested that HIP1 might offer a valuable therapeutic target.
Integrative analysis of mRNA and miRNA interaction between high and low HIP1 expressers
We analyzed the gene expression patterns of leukemia blasts from 131 patients with low expression and 66 patients with high expression from the published TCGA data15. We found 475 genes were down-regulated and 662 genes were up-regulated in high expressers (Figure S5). By means of miRNA-mRNA integrative analysis, we found several targeted genes of miR-28-5p, miR-15a, miR-16 and miR-660. Specifically, among these 1137 aberrantly expressed genes, 84 genes were predicted to be targeted by miR-28-5p, 100 by miR-15a, 100 by miR-16 and 58 by miR-600 (Figure S6–9). Notably, these targeted genes were involved in different regulatory networks. In the KEGG analysis, these targeted genes of miR-28-5p, miR-15a and miR-16, miR-660 respectively involved in 77, 70, 83 and 33 different metabolic networks with oncogenic potential (Table S7–10).
Discussion
In this study, we uncover high HIP1 expression could predict unfavorable overall survival in AML patients. Additionally, we found distinct microRNA signatures associated with high HIP1 expression in AML. These results were also validated in an independent cohort of AML patients. Thus, we provide sound evidence that HIP1 expression analysis can add to risk classification and therapy decision making for AML patients.
The HIP1 gene is located on chromosome 7q11.23 and encodes a 116-kDa protein6. This protein can interact with clathrin, actin, and inositol lipid and involve in receptor trafficking, including regulating cell surface expression of receptor tyrosine kinases6. The activated tyrosine kinase signal is important for leukemogenesis. Frequent mutations of tyrosine kinase genes like FLT3, KIT, NRAS and JAK2 in de novo AML were well documented. In our study, we found HIP1 expressers had a higher frequency of FLT3-ITD positive in our cohort, although the difference is not significant. By contrast, in patients from the TCGA cohort, high HIP1 expression was positively correlated with FLT3-ITD positive. The discrepancy might be attributed to the lower frequency of FLT3-ITD in our cohort than in the Western cohort. We also found patients with high expression of HIP1 had lower levels of hemoglobin. One possible reason may be the hypothesis that overexpression of HIP1 in blasts can stabilize or even increase levels of transferrin receptor as reported9 and in turn promote utilization of iron for blasts, leading to iron deficiency in normal red blood cells. Interestingly, HIP1 overexpression enables prostate cancer cells to metastasis through increasing the expression of integrin6. The similarities of high HIP1 expression between FAB subtype M4/M5 blasts and the prostate cancer cells suggest an analogous promoting metastasis role for HIP1 through regulations of cytoskeletal processes and integrin expression. Analogously, HIP1 expression might facilitate M4/M5 blasts to migrate into extramedullary organs. What is very interesting and consistent with our results is that Roel G. W. et al. show NPM1 mutant AML blasts have higher level of HIP1 expression by gene microarray analysis16. It is conceivable that high HIP1 expressers will associate with DNMT3A mutations. The reason might be that both DNMT3A mutations and HIP1 overexpression are predominant in FAB M4/M5 subtype patients. However, the reason why HIP1 expression is negatively associated with CEBPA double allele mutation in our cohort but not in TCGA cohort is unclear. As mentioned above, these results implied AML cases with high HIP1 expression might be more resistant to chemotherapy, and associated with a poorer outcome. In this study, we find that high HIP1 expressers harbored poor overall survival in two different cohorts. This is consistent with the report that HIP1 overexpression with oncogenic property is an independent predictor of relapse in patients with prostate cancer17. By contrast, Hsu et al. reported HIP1 functions as a potential tumor suppressor18. They observed that reduced expression of HIP1 in lung adenocarcinoma cells leads to development of late metastases and poor prognosis. Taken together, these conflicting data in solid tumors indicate that functions of HIP1 need much more experimental clarification.
In order to further understand the biologic insight of aberrant HIP1 expression, we conducted the miRNAs analysis in AMLs. Among differentially expressed miRNAs, we found 31 miRNAs had dysregulated expression in our patients. Among the 18 upregulated expression of miRNAs, 4 miRNAs including miR-15a, miR-16-2, miR-28 and miR-660 were validated in a large cohort of patients. More importantly, these 4 miRNAs were also downregulated after silencing HIP1 expression in both Kasumi-1 and THP-1 cell lines. These results implied that one of the main mechanisms of HIP1 in the oncogenic propensity might directively or indirectively act through these miRNAs. In order to understand the biological insight of these miRNAs, miRNA-mRNA interaction were carried out in silico analyses. Consequently, these miRNAs could affect 342 out of 1137 (30%) genes that significantly changed between high and low HIP1 expressers in TCGA data set. These targeted genes were involving in 263 metabolic pathways in KEGG pathway analysis. The miRNAs were functionly involved in several important pathways. For example, CCND3 gene regulated by miR-28-5p involved in P53 pathway, Wnt signaling pathway, cell cycle and Jak-STAT signaling pathway (Table S7), several targeted genes (ZYX, VCL, PDPK1, MAPK9, COL1A1, Tables S8 and 9) of miR-15/16 were involved in adhesion or migration processes; LFNG in notch signaling pathway was regulated by miR-660 (Table S10), etc. Moreover, these miRNAs have been proved to be important prognostic markers and novel targets for therapy in cancers. Although miR-15 and miR-16 are mainly reported to be tumor suppressors, they have been reported to be upregulated in various kinds of cancer and be correlated with tumor cells metastasis, indicating their potential roles as oncomiRs5. miR-660 expression was used as a good candidate for prognosis prediction in breast cancer19. In addition, increased miR-28 expression leads to autonomous growth of hematopoietic cells by constitutive activation of STAT520. These differentially expressed microRNAs may help us further understand the biologic insights of poor survival in patients with high HIP1 expression.
There are still some limitations in this study. Firstly, we only examine genes of FLT3-ITD, NPM1, CEBPA and DNMT3A mutations, thus we could not exclude other genes like IDH1/2, TET2, ASXL1 mutations those will confound the prognostic value of HIP1 expression in AML patients. Secondly, the putative interaction of miRNA and mRNA uncover several important regulatory networks, but luciferase reporter assays are required to further study in the future. Finally, functional study is limited to the silencing HIP1 expression on proliferation in leukemia cell lines in vitro, enforced expression of HIP1 and in vivo models are also required to investigate the oncogenesis of HIP1. Therefore, caution in application of our findings is still warranted.
In conclusion, we present high HIP1 expression as a reliable and powerful prognostic factor for AML.
Materials and Methods
Patients
Clinical data were abstracted from medical records of AML patients in Zhejiang Institute of Hematology (ZIH), China. Between January 2010 and July 2015, 270 patients with detailed diagnoses and treatment information were included in this study. WHO classification, conventional cytogenetic banding assay, and molecular analyses were performed as previously described in AML diagnosis21. Cytogenetic groups of patients were classified as favorable, intermediate, and unfavorable risk according to the NCCN guideline22. Favorable subgroups included t(8; 21)/AML1-ETO and inv16/CBFβ-MYH11; adverse consisted of t(9; 22), inv(3)/t(3; 3), −5, −7, del(5q), del(7p), 11q23 and complex translocations; intermediate subgroups contained cytogenetically normal (CN) and AML with other cytogenetic abnormalities. CN -AML was defined as AML with the karyotype 46, XY [20] or 46 XX [20] in all 20 metaphase cells analyzed. Patients were treated with intensive induction chemotherapy as previous reported23,24. In the consolidation therapy, younger patients were treated with a high-dose cytarabine-based chemotherapy23. The chemotherapy consolidation for elderly patients was decided by the physicians in an individualized manner, as described previously23. No patient in our study received allogeneic transplantation. Patients with secondary AML or acute promyelocytic leukemia were excluded. This study was approved by the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (No. 2016313). Written informed consent was obtained from all participating subjects. All the study methods were carried out in accordance with the approved guidelines.
Cytogenetic and Gene mutation analysis
The bone marrow (BM) samples of de novo AML patients were analyzed by R-banding analysis. Chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature25. DNA and RNA samples of AML patients were obtained from mononuclear cells isolated by Ficoll gradient centrifugation from bone marrow samples at primary diagnosis. Gene mutations of NPM1, FLT3-ITD, and CEBPA were analyzed by whole-gene sequencing as previously described26. RNA samples were used to determine PML-RARA, AML1-ETO, and CBFβ-MYH11 fusion genes by reverse transcription polymerase chain reaction (RT-PCR).
Quantitative reverse transcriptase-PCR
RNA was extracted using RNeasy Mini kit (Qiagen, Venlo, Netherlands) and first-strand complementary DNA synthesis was performed using the MMLV systems (Life Technologies). Quantitative PCR was performed in triplicate using SYBR-Green PCR Master Mix kit (Takara, Japan) on an IQ5 real time PCR instrument (Bio-Rad, USA), using standard settings: 95 °C (1 min), 40 cycles of 95 °C (5 s) and 60 °C (1 min). mRNA levels were normalized to GAPDH housekeeping gene. The following primers were used for quantitative PCR: HIP1 5′-GCGGCTCATTCAGATCCCC-3′ (sense) and 5′-GAGGTCATCCTTCTCTAGGACTG-3′ (antisense); GAPDH (control), 5′-ATGGGGAAGGTGAAGGTCG-3′ (sense) and 5′-GGGTCATTGATGGCAACAATATC-3′ (antisense). PCR reactions were performed in a total volume of 25 μl containing of 1 μl of 100 ng/μl sample cDNA, 12.5 μl of 2 × PCR Mix, 1 μl of 0.5 μM of each primer, and 10.5 μl of ddH2O.
MicroRNA experiments
For the miRNA profiling, total RNA was extracted and purified using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, US) following the manufacturer’s instructions. RNA integrity number (RIN) was assessed by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US). miRNA expression was performed using the Agilent Human miRNA Microarray Kit Version 16.0. Total RNA (100 ng) was hybridized per sample and processed according to the manufacturer’s instructions. The arrays were scanned by an Agilent Technology G2565BA scanner. The scanned images were gridded and analyzed with Agilent Feature Extraction Software Version 10.7. Raw data were normalized by quantile algorithm using Gene Spring Software 11.0. Each microRNA signature was represented by the average of its expression value of replicate probes.
Cell culture and transfections
Kasumi-1 and THP-1 cell lines were purchased from a typical cell culture collection Committee of the Chinese Academy of Sciences Library. Cells were cultured in RPMI 1640 medium (Corning, USA) containing 10% fetal bovine serum. Cells were maintained at 37 °C, 5% CO2, 95% air and 100% relative humidity. The HIP1 siRNA and negative control were purchased from Vigene Biosciences (Shangdong, China). HIP1 siRNA and negative control were transfected in THP-1 and Kasumi-1 cells using protocol provided by the manufacturer. To monitor the effect of siRNA on gene silencing, 5 × 105 cells were seeded in 6-well plates in 2 mL medium containing 5 μl LipofectamineTM3000 transfection reagent and 50 nM siRNA for 72 hours. Down-regulation of HIP1 expression was detected by quantitative real-time PCR analysis. The oligo sequences are available in the supplementary methods.
Cell proliferation assay
Cell proliferation assays were performed in triplicate with THP-1 and Kasumi-1 cells transfected with and without the HIP1-siRNA and negative control (NC) siRNA. The experiment was subdivided into four groups: HIP1 siRNA, NC siRNA, blank control and culture only. The treated cells with only lipofectamine reagent were considered as a blank control. Specifically, 100 ul of cells (5 × 105 cells/ml) were plated into 96-well plates. 10 ul of CellTiter 96 Aqueous One Solution Cell Proliferation Assay solution (Promega, USA) was added to each well at post transfection 24, 48 and 72 hours. Plates were read in 490 nm. Growth curves were generated by quantifying the relative number of viable cells.
Expression of microRNAs after silencing HIP1 expression
After 72 hours, total RNA was isolated from transfected cells. qTR-PCR was performed using ALL-in-one miRNA real-time quantitative reverse transcription PCR (qRT-PCR) detection kit (GeneCopeia,USA). Total RNA (2 μg) was incubated with miRNA reverse transcription (RT) reagents at 37 °C for 60 min, 85 °C 5 mim in a total volume of 25 μl. The cDNA product was stored −20 °C until being analyzed with RT-PCR. To detect the miRNAs, 2 μl cDNA product was amplified using 2 μl miRNA qPCR primer (2 μM), 2 μl the universal adaptor PCR primer (2 μM) and 10 μl ALL-in-one PCR mix in 20 μl PCR System. The reactions were incubated in a 96-well plate at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 sec, 58 °C for 20 sec and 72 °C for 10 sec. All sample were run in triplicate. The relative quantification of the target gene expression was determined using the 2−ΔΔCT method and U6 was used for normalization. All primers were seen in the Supplementary Methods.
Definition of clinical end points and statistical analysis
Patient characteristics were summarized using descriptive statistics, which included frequency counts, median, and range. The main objective of this study was to evaluate the prognostic impacts of HIP1 expression on Overall survival (OS) of AML patients. OS was defined as time from date of diagnosis until death due to any cause or the last follow-up. We used AML cohort from TCGA (https://tcga-data.nci.nih.gov/tcga/) as a validation cohort, which contains publicly available data of gene microarray expression and clinical information. Determination of optimal cutoff value for HIP1 expression in our study and the validation TCGA cohort was done with Cutoff Finder using log-rank test (http://molpath.charite.de/cutoff/). To test the robustness of HIP1 gene as a prognostic biomarker, the resampling statistics of the multiple survival screening (MSS) algorithm was used as reported by Jie Li et al.14. First, we used the TCGA dataset to generate the survival genes with the “samr” R package27. Second, we generated 36 random datasets (RDSs) with the 44 of 131 high HIP1 expressers and 22 of 66 low HIP1 expressers in the TCGA cohort. Additionally, we generate 10000 random gene sets (RGSs) each containing 30 genes which were randomly selected from the 45 survival genes. For each RGS-RDS pair, we used the R-code of MSS as reported by Jie Li et al.14 to calculate the survival screening P-value of the RGS and identify the top 30 genes signatures. The proportional-hazards assumption was checked for each variable before fitting Cox models. Variables with a p-value < 0.2 and the well-established predictors were selected as adjustment covariates into the multivariable analyses. A nonparameter T-test was used to test for the difference of microRNA signatures between high and low HIP1 expressers. Hierarchical clustering based on expression levels of these microRNAs was performed and visualized by heatmap. Interaction of miRNA and mRNA integrative analyses in silico were using the mirtar platform (http://mirtar.mbc.nctu.edu.tw/human/index.php). All statistical analyses were conducted with R statistic packages, version 2.15.0 (www.r-project.org). The two-sided level of significance was set at p-value < 0.05.
Additional Information
How to cite this article: Wang, J. et al. Prognostic significance of huntingtin interacting protein 1 expression on patients with acute myeloid leukemia. Sci. Rep. 7, 45960; doi: 10.1038/srep45960 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Khaled, S., Al Malki, M. & Marcucci, G. Acute Myeloid Leukemia: Biologic, Prognostic, and Therapeutic Insights. Oncology 30, 318–329 (2016).
Papaemmanuil, E. et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. The New England journal of medicine 374, 2209–2221, doi: 10.1056/NEJMoa1516192 (2016).
Goswami, C. P. & Nakshatri, H. PROGgene: gene expression based survival analysis web application for multiple cancers. Journal of clinical bioinformatics 3, 22, doi: 10.1186/2043-9113-3-22 (2013).
Stirewalt, D. L. et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes, chromosomes & cancer 47, 8–20, doi: 10.1002/gcc.20500 (2008).
Huang, E., Liu, R. & Chu, Y. miRNA-15a/16: as tumor suppressors and more. Future oncology 11, 2351–2363, doi: 10.2217/fon.15.101 (2015).
Hyun, T. S. & Ross, T. S. HIP1: trafficking roles and regulation of tumorigenesis. Trends in molecular medicine 10, 194–199, doi: 10.1016/j.molmed.2004.02.003 (2004).
Rao, D. S. et al. Huntingtin interacting protein 1 Is a clathrin coat binding protein required for differentiation of late spermatogenic progenitors. Molecular and cellular biology 21, 7796–7806, doi: 10.1128/MCB.21.22.7796-7806.2001 (2001).
Ames, H. M. et al. Huntingtin-interacting protein 1 phosphorylation by receptor tyrosine kinases. Molecular and cellular biology 33, 3580–3593, doi: 10.1128/MCB.00473-13 (2013).
Rao, D. S. et al. Altered receptor trafficking in Huntingtin Interacting Protein 1-transformed cells. Cancer cell 3, 471–482 (2003).
Bradley, S. V. et al. Huntingtin interacting protein 1 is a novel brain tumor marker that associates with epidermal growth factor receptor. Cancer research 67, 3609–3615, doi: 10.1158/0008-5472.CAN-06-4803 (2007).
Marghalani, S., Feller, J. K., Mahalingam, M. & Mirzabeigi, M. Huntingtin interacting protein 1 as a histopathologic adjunct in the diagnosis of Merkel cell carcinoma. International journal of dermatology 54, 640–647, doi: 10.1111/ijd.12454 (2015).
Bradley, S. V. et al. Aberrant Huntingtin interacting protein 1 in lymphoid malignancies. Cancer research 67, 8923–8931, doi: 10.1158/0008-5472.CAN-07-2153 (2007).
Ross, T. S., Bernard, O. A., Berger, R. & Gilliland, D. G. Fusion of Huntingtin interacting protein 1 to platelet-derived growth factor beta receptor (PDGFbetaR) in chronic myelomonocytic leukemia with t(5;7)(q33;q11.2). Blood 91, 4419–4426 (1998).
Li, J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nature communications 1, 34, doi: 10.1038/ncomms1033 (2010).
Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine 368, 2059–2074, doi: 10.1056/NEJMoa1301689 (2013).
Verhaak, R. G. et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 106, 3747–3754, doi: 10.1182/blood-2005-05-2168 (2005).
Rao, D. S. et al. Huntingtin-interacting protein 1 is overexpressed in prostate and colon cancer and is critical for cellular survival. The Journal of clinical investigation 110, 351–360, doi: 10.1172/JCI15529 (2002).
Hsu, C. Y. et al. Huntingtin-Interacting Protein-1 Is an Early-Stage Prognostic Biomarker of Lung Adenocarcinoma and Suppresses Metastasis via Akt-mediated Epithelial-Mesenchymal Transition. American journal of respiratory and critical care medicine 193, 869–880, doi: 10.1164/rccm.201412-2226OC (2016).
Krishnan, P. et al. Next generation sequencing profiling identifies miR-574-3p and miR-660-5p as potential novel prognostic markers for breast cancer. BMC genomics 16, 735, doi: 10.1186/s12864-015-1899-0 (2015).
Girardot, M. et al. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood 116, 437–445, doi: 10.1182/blood-2008-06-165985 (2010).
Wang, J. H. et al. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci USA 110, 17017–17022, doi: 10.1073/pnas.1315558110 1315558110 (2013).
O’Donnell, M. R. et al. Acute myeloid leukemia. J Natl Compr Canc Netw 10, 984–1021, doi: 10/8/984 (2012).
Ma, Q. L. et al. High IDH1 expression is associated with a poor prognosis in cytogenetically normal acute myeloid leukemia. Int J Cancer, doi: 10.1002/ijc.29395 (2014).
Jin, J. et al. Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: a multicentre, open-label, randomised, controlled phase 3 trial. The Lancet. Oncology 14, 599–608, doi: 10.1016/S1470-2045(13)70152-9 (2013).
Brothman, A. R., Persons, D. L. & Shaffer, L. G. Nomenclature evolution: Changes in the ISCN from the 2005 to the 2009 edition. Cytogenet Genome Res 127, 1–4, doi: 10.1159/000279442000279442 (2009).
Chen, W. L. et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood 124, 1645–1654, doi: 10.1182/blood-2014-02-554204blood-2014-02-554204 (2014).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America 98, 5116–5121, doi: 10.1073/pnas.091062498 (2001).
Acknowledgements
We are very thankful to Ms. Jennifer Strong from University of Cincinnati College of Medicine for carefully reading and polishing the manuscript. We would like to thank the patients who donated leukemia specimens. This work was supported by National Natural Science Foundation of China grants (81370643, 81470305, U1404806), National Public Health Grand Research Foundation (201202017) and the Foundation of Innovation Team for Basic and Clinical Research of Zhejiang Province (2011R50015).
Author information
Authors and Affiliations
Contributions
J.H.W. and J.J. designed the research and/or analyzed the data. M.X.Y., Q.G., Q.L.M., C.H., Z.X.M., X.F.Y., X.L., Y.G.W., H.Z.P., D.M.W., J.S.H. carried out the molecular genetic studies and in vitro experiments of leukemia cell line models, H.T.M., H.Y.T., W.B.Q., and J.J. provided clinical data. J.H.W. and J.J. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, J., Yu, M., Guo, Q. et al. Prognostic significance of huntingtin interacting protein 1 expression on patients with acute myeloid leukemia. Sci Rep 7, 45960 (2017). https://doi.org/10.1038/srep45960
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep45960
This article is cited by
-
Transformation of fibroblast‐like synoviocytes in rheumatoid arthritis; from a friend to foe
Autoimmunity Highlights (2021)
-
Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.