Abstract
Streptococcus pneumoniae can be classified in more than 90 capsular types, as traditionally determined by serological methods and more recently by PCR-based techniques. Such methods, however, can be expensive, laborious or unable to accurately discriminate among certain serotypes. Therefore, determination of capsular types, although extremely important for epidemiological purposes and for estimating the impact of pneumococcal conjugate vaccines, is mainly restricted to research laboratories, being rarely performed in the clinical setting. In the present study, MALDI-TOF MS was evaluated as an alternative tool to characterize 416 pneumococcal isolates belonging to serotypes 6A, 6B, 6C, 9N, 9V or 14. For MALDI-TOF MS analysis, each isolate was submitted to an extraction protocol using formic acid and acetonitrile. Measurements were performed with a Bruker Microflex LT mass spectrometer using default parameters and generating spectra in the range of 2,000–20,000 m/z. Spectra were analyzed with the BioNumerics software v7.6. Isolates were mainly distributed according to the capsular type in a Neighbor Joining tree and serotypes investigated were successfully discriminated by the presence/absence of 14 selected biomarkers. The results suggest that MALDI-TOF MS is a promising alternative for typing pneumococcal strains, highlighting its usefulness for rapid and cost-effective routine application in clinical laboratories.
Similar content being viewed by others
Introduction
Streptococcus pneumoniae is a major human pathogen that can be also found as nasopharyngeal colonizers among variable numbers of asymptomatic individuals worldwide1. The polysaccharide capsule is the basis for characterizing pneumococcal isolates into serotypes, and represents the target of pneumococcal conjugate vaccines (PCV) currently available2. Although more than 90 capsular types have been reported, a restricted number of serotypes stands out for their high prevalence in both pneumococcal disease and nasopharyngeal colonization, as well as for association with antimicrobial resistance. Serotype 14 and serotypes comprised by serogroups 6 and 9 are important examples, and most of them have already been included in PCVs2,3,4. In Brazil, the first PCV introduced in the National Immunization Program for routine free of charge childhood vaccination was the 10-valent vaccine (PCV10) in 2010, comprising serotypes 1, 4, 5, 6B, 7F, 9V, 14, 18C, 19F, and 23F5. Simultaneously, the 13-valent vaccine (PCV13), containing the additional serotypes 3, 6A, and 19A, was made commercially available in private clinics.
Surveillance of pneumococcal serotypes is important, among other purposes, to evaluate vaccination effects. The standard methodology for capsular typing is the Quellung reaction, which is mostly limited to reference centers due to the strict requirements regarding preparation and quality control of antisera and training of the staff involved. Latex agglutination, PCR amplification or sequencing of capsular genes represent some of the alternative approaches2. These methods, however, can be expensive and laborious; and some of them do not accurately discriminate among serotypes within a same serogroup, such as serogroup 6. Hence, determination of capsular types can be restricted, especially in laboratories of resource-limited countries, being rarely performed in the clinical setting.
MALDI-TOF MS is an emerging technology that has been increasingly used in clinical laboratories for rapid bacterial identification6. Moreover, it has also showed the potential for typing applications among streptococcal species7,8,9,10. In the present study, MALDI-TOF MS was evaluated as an alternative tool to characterize Streptococcus pneumoniae isolates according to the capsular type.
Methods
A total of 416 Streptococcus pneumoniae isolates were analyzed, including 141 strains of serotype 14, 49 strains of serotype 6A, 143 strains of serotype 6B, 38 strains of serotype 6C, 24 strains of serotype 9N and 21 strains of serotype 9V.
All the isolates were recovered during surveillance studies performed by our group or were received from various national health institutions for species confirmation and capsular typing, between 1988 and 2011. Isolates obtained from diseased individuals, including children and adults, were recovered from clinical specimens taken as part of standard patient care procedures and did not require ethical approval for their use. Isolates from carriage studies, including both children and adults, were recovered from clinical specimens as approved by the ethics committees of the institutions involved. The isolates were previously identified by phenotypic tests, including observation of colony morphology and type of hemolysis on blood agar plates; cellular characteristics after Gram stain; optochin susceptibility and bile-solubility11. The capsular types were previously determined for all 416 strains by the Quellung reaction with antisera kindly provided by the Streptococcus Laboratory at the Centers for Disease Control and Prevention (CDC, GA, USA), and also by multiplex PCR for a fraction of the isolates12,13,14,15,16,17.
For MALDI-TOF MS analysis, a quick extraction protocol was performed for all isolates as follows. After overnight growth, five bacterial colonies were suspended in 5 μl of formic acid 70% (Tedia, Cincinatti, OH, USA), which was then vigorously homogenized during 10 s. Subsequently, 5 μl of acetonitrile (Tedia) were added to the suspension, which was then gently homogenized. The suspension was centrifuged at 5000 rpm for 3 min, and 1 μl of the supernatant was poured in a spot of the target plate (MSP 96 target polished steel BC, Bruker Daltonics, Germany). After drying, each spot was covered with 1 μl of CHCA matrix (α-Cyano-4-hydroxycinnamic acid; Bruker Daltonics).
Measurements were performed with the Microflex LT mass spectrometer (Bruker Daltonics) and Biotyper software using the default parameters (laser frequency of 60 Hz, ion source voltages of 2.0 and 1.8 kV, and lens voltage of 6 kV), which generated spectra in the range of 2,000–20,000 m/z. Spectra were then exported to the BioNumerics software v7.6 (Applied Maths, Ghent, Belgium), where the raw spectra were preprocessed and normalized using default parameters (baseline subtraction was performed with Running Rolling disc, and peak detection was performed using a signal to noise ratio of 10). After preprocessing, spectra were submitted to peak matching using a tolerance of ±0,002 m/z.
All biomarkers automatically detected by BioNumerics were exported to a spreadsheet containing all strains (see Supplementary Spreadsheet S1). This spreadsheet was visually analyzed by counting the number and calculating the percentage of strains containing a certain biomarker within each serotype. After analysis, a set of biomarkers was selected to constitute serotype-specific profiles.
In addition, a Neighbor Joining tree based on Pearson coefficient was constructed using BioNumerics.
Diversity of serotypes and MALDI profiles was assessed by the Simpson’s Index of Diversity (SID)18, and congruence between serotype and MALDI profile was estimated by the Adjusted Wallace Coefficient (AW), with 95% confidence intervals (95% CI)19. SID and AW were calculated using the online tool available at http://www.comparingpartitions.info.
Reference or internal control S. pneumoniae strains (strain ATCC700902 of serotype 14, strain Sp1019 of serotype 9N, strain ATCC700671 of serotype 9V, strain ATCCBAA659 of serotype 6A, strain ATCC700675 of serotype 6B and strain 2008008385 of serotype 6C) were also analyzed.
Results and Discussion
In the Neighbor Joining tree, isolates were distributed mainly according to the capsular type (Fig. 1), and 12 clusters were detected, including 10 major and two minor clusters. Distribution of isolates according to serotype and major MALDI clusters is shown in Table 1.
Isolates belonging to certain capsular types, such as 9N and 9V, showed a more random distribution across the tree, while strains belonging to other serotypes, such as 6B and 14, showed a tendency to a more homogeneous clustering. Interestingly, capsular types 9N and 9V were the serotypes less represented in the study, comprising around 20 isolates each. In turn, serotypes 14 and 6B included the largest numbers of strains evaluated in the present study (around 140 each), suggesting that clustering might be enhanced as more strains are included in the analysis.
Congruence between MALDI-TOF cluster and capsular type was observed (AW = 0.604; 95% CI 0.527–0.681). In addition, a higher diversity (SID = 0.863) was observed regarding MALDI profiles when compared to serotype distribution (SID = 0.742). These observations indicate that MALDI-TOF MS results are congruent with capsular typing, and are also more discriminatory, suggesting that this technique might be useful to predict intra-serotype variations, such as those related to distribution of clones and antimicrobial susceptibility profiles.
In general, isolates belonging to serotypes 9N and 9V were mainly clustered together with serotype 14 isolates. Likewise, serotype 6A strains were mostly allocated together with serotype 6C isolates. These results corroborate the close genetic relationship between serotype 14 and serogroup 920, as well as between serotypes 6A and 6C21,22, suggesting that MALDI-TOF MS also has potential for inferring phylogenetic relationship.
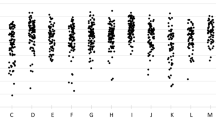
A total of 35 peaks (or biomarkers) were detected, and they ranged from 2,633 to 10,405 m/z (see Supplementary Spreadsheet S1). After analysis of these biomarkers, 14 were selected to compose serotype-specific profiles (Fig. 2). The profiles (presence/absence) of selected biomarkers were successful for discriminating all serotypes evaluated, even those that presented a more scattered distribution in the Neighbor Joining tree, such as 9N and 9V (Fig. 1).
*Position in the spectra using a tolerance of ± 0,002 m/z. Green indicates that the biomarker is present in more than 90% of the strains of a given serotype; Yellow, present in 70–89% of the strains; Light red, present in 50–69% of the strains; Red, absent in all strains. The sets of 3 biomarkers useful to discriminate among serotypes within serogroups 6 and 9 are highlighted in blue and brown, respectively.
Selected biomarkers were also classified according to their distribution within each serotype, as follows: green when present in more than 90% of the strains of a given serotype; yellow when present in 70–89% of the strains; light red when present in 50–69% of the strains; and red when absent in all strains of a given serotype. To perform a preliminary validation, these biomarkers were investigated in reference or internal control S. pneumoniae strains of each serotype evaluated and all of them were shown to possess the expected profile.
Additionally, differentiation among serotypes within a single serogroup (as in serogroups 6 and 9) was achieved by the analysis of sets of 3 biomarkers (Fig. 2). Serogroup 9 comprises serotypes 9A, 9L, 9N, and 9V; likewise, serogroup 6 includes serotype 6A, 6B, 6C, and 6D2. Serotypes 9A, 9L, and 6D are still rarely detected13,15. In our collection, only three strains belonged to serotype 9A, two to serotype 9L and only one to serotype 6D. Therefore, due to the lack of representativeness, strains belonging to such serotypes were not included in the main analysis. However, the profile of the 3-biomarker sets used to differentiate serotypes within serogroups 6 and 9 was also evaluated in those strains and they indeed exhibited different profiles, being distinguished from the other serotypes in the same serogroup. Although a higher number of isolates belonging to those less commonly reported serotypes should be evaluated, our results suggest that MALDI-TOF MS could work as a complementary methodology to determine serotype once serogroup has been established by other means, such as PCR.
Determination of pneumococcal capsular types is essential for different purposes. Specifically, discrimination among serotypes comprised by the same serogroup is crucial since some of them are included in PCVs (such as serotypes 6A and 6B of serogroup 6 and serotype 9V of serogroup 9). On the other hand, serotypes 6C and 9N are not present in any of the available conjugate vaccines and, thus, have the potential to emerge in this scenario2. Indeed, the emergence of serotype 6C has been detected in the USA after PCV7 implementation14,15.
Therefore, a more suitable and accessible methodology to determine the capsular type of pneumococcal isolates is needed. Recently, MALDI-TOF MS was evaluated for this purpose, showing the potential for differentiating ten major pneumococcal serotypes in Japan, including serotypes 3, 6B, 15A, 15C, 19A, 19F, 23A, 24F, 35B, and 387. However, the ability to distinguish the various serotypes comprised by serogroup 6, as well as to differentiate serotype 14 and those within serogroup 9 investigated in the present study, has not been previously evaluated.
This work contributes for the development and evaluation of alternative methodologies for the capsular typing of pneumococcal strains, indicating that MALDI-TOF MS is a promising tool. Nevertheless, as every novel and recently proposed scientific approach, it needs external validation in order to assess the reproducibility of results among pneumococcal strains recovered from other places of the world. Still, the observation that a quick and simplified extraction protocol was sufficient to generate high quality MALDI-TOF spectra highlights its usefulness for rapid and cost-effective routine application in clinical and resource-limited laboratories.
Additional Information
How to cite this article: Pinto, T. C. A. et al. Potential of MALDI-TOF MS as an alternative approach for capsular typing Streptococcus pneumoniae isolates. Sci. Rep. 7, 45572; doi: 10.1038/srep45572 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Weiser, J. N. The pneumococcus: why a commensal misbehaves. J Mol Med (Berl) 88, 97–102 (2010).
Geno, K. A. et al. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28, 871–899 (2015).
Dagan, R. Antibiotic resistance and the potential impact of pneumococcal conjugate vaccines. Commun Dis Intell Q Rep 27, 134–142 (2013).
Camargo, P. et al. Penicillin resistance and serotyping of Streptococcus pneumoniae in Latin America. Paediatr Respir Rev 7, 209–214 (2006).
Ministério da Saúde. Proposta para a introdução da vacina pneumocócica 10-valente (conjugada) no calendário básico de vacinação da criança. Brasília (2010).
Clark, A. E. et al. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26, 547–603 (2013).
Nakano, S. et al. Development and evaluation of MALDI-TOF MS-based serotyping for Streptococcus pneumoniae . Eur J Clin Microbiol Infect Dis 34, 2191–2198 (2015).
Williamson, Y. M. et al. Differentiation of Streptococcus pneumoniae conjunctivitis outbreak isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 74, 5891–5897 (2008).
Moura, H. et al. MALDI-TOF mass spectrometry as a tool for differentiation of invasive and noninvasive Streptococcus pyogenes isolates. FEMS Immunol Med Microbiol 53, 333–342 (2008).
Lartigue, M. F. et al. Rapid detection of “highly virulent” Group B Streptococcus ST-17 and emerging ST-1 clones by MALDI-TOF mass spectrometry. J Microbiol Methods 86, 262–265 (2011).
Spellerberg, B. & Brandt, C. In Manual of Clinical Microbiology Vol. 1: Streptococcus (eds Jorgensen, J. H. et al.) 383–402 (American Society for Microbiology, 2015).
Dias, C. et al. Sequential multiplex PCR for determining capsular serotypes of pneumococci recovered from Brazilian children. J Med Microbiol 56, 1185–1188 (2007).
Pinto, T. C. et al. Streptococcus pneumoniae of serotypes 9 and 14 circulating in Brazil over a 23-year period prior to the introduction of PCV10: role of international clones in the evolution of antimicrobial resistance and description of a novel genotype. Antimicrob Agents Chemother 60, 6664–6672 (2016).
Neves, F. P. et al. Nasopharyngeal carriage, serotype distribution and antimicrobial resistance of Streptococcus pneumoniae among children from Brazil before the introduction of the 10-valent conjugate vaccine. BMC Infect Dis. 13, 318–325 (2013).
Sessegolo, J. F. et al. Distribution of serotypes and antimicrobial resistance of Streptococcus pneumoniae strains isolated in Brazil from 1988 to 1992. J Clin Microbiol 32, 905–911 (1994).
Teixeira, L. M. et al. Serotyping distribution and antimicrobial resistance of Streptococcus pneumoniae isolated in Brazil (1992–1996). Adv Exp Med Biol 418, 269–271 (1997).
Mendonça-Souza, C. R. et al. Occurrence and characteristics of erythromycin-resistant Streptococcus pneumoniae strains isolated in three major Brazilian states. Microb Drug Resist 10, 313–320 (2004).
Hunter, P. R. & Gaston, M. A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol 26, 2465–2466 (1988).
Severiano A. et al. Adjusted Wallace Coefficient as a Measure of Congruence between Typing Methods. J Clin Microbiol 49, 3997–4000 (2011).
Coffey, T. J. et al. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiol 145, 2023–2031 (1999).
Carvalho, M. da G. et al. Active Bacterial Core Surveillance Team. PCR-based quantitation and clonal diversity of the current prevalent invasive serogroup 6 pneumococcal serotype, 6C, in the United States in 1999 and 2006 to 2007. J Clin Microbiol 47, 554–559 (2009).
McEllistrem, M. C. & Nahm, M. H. Novel pneumococcal serotypes 6C and 6D: anomaly or harbinger. Clin Infect Dis 55, 1379–1386 (2012).
Acknowledgements
We thank Dr Lesley McGee for the kind donation of pneumococcal reference strains; Dr Hercules Moura for the valuable help with MALDI-TOF MS issues; and Filomena Soares Pereira da Rocha and Jaqueline Martins Morais for technical support. This work was supported in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
Author information
Authors and Affiliations
Contributions
T.C.A.P., J.M.P. and L.M.T. conceived and designed the study and wrote the manuscript. T.C.A.P. analyzed the data and performed statistical analyses. N.S.C., L.F.S.C., R.L.R., A.C.N.B. and F.P.G.N. performed the conventional characterization of the isolates and the MALDI-TOF MS runs. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Pinto, T., Costa, N., Castro, L. et al. Potential of MALDI-TOF MS as an alternative approach for capsular typing Streptococcus pneumoniae isolates. Sci Rep 7, 45572 (2017). https://doi.org/10.1038/srep45572
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep45572
This article is cited by
-
MALDI-TOF mass spectrometry for sub-typing of Streptococcus pneumoniae
BMC Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.