Abstract
The cytochrome P450 enzyme OleTJE from Jeotgalicoccus sp. ATCC 8456 is capable of converting free long-chain fatty acids into α-alkenes via one-step oxidative decarboxylation in presence of H2O2 as cofactor or using redox partner systems. This enzyme has attracted much attention due to its intriguing but unclear catalytic mechanism and potential application in biofuel production. Here, we investigated the functionality of a select group of residues (Arg245, Cys365, His85, and Ile170) in the active site of OleTJE through extensive mutagenesis analysis. The key roles of these residues for catalytic activity and reaction type selectivity were identified. In addition, a range of heterologous redox partners were found to be able to efficiently support the decarboxylation activity of OleTJE. The best combination turned out to be SeFdx-6 (ferredoxin) from Synechococcus elongatus PCC 7942 and CgFdR-2 (ferredoxin reductase) from Corynebacterium glutamicum ATCC 13032, which gave the highest myristic acid conversion rate of 94.4%. Moreover, Michaelis-Menton kinetic parameters of OleTJE towards myristic acid were determined.
Similar content being viewed by others
Introduction
Cytochrome P450 (CYP) enzymes broadly existing in archaea, prokaryotes, and eukaryotes belong to the ubiquitous superfamily composed of diverse functional oxygenases1,2. These versatile biocatalysts are capable of mediating a great variety of natural and unnatural reactions2,3,4,5,6. Mechanistically, P450 enzymes can be divided into monooxygenases, peroxidases, and peroxygenases based on their catalytic properties2.
P450 monooxygenases utilize O2 as oxygen donor and two electrons transferred from NAD(P)H by redox partner protein(s) to the heme iron reactive center, to catalyze the monooxygenation of numerous substrates3,7. By contrast, P450 peroxygenases, such as P450 OleTJE from Jeotgalicoccus sp. ATCC 8456, P450BSβ from Bacillus subtilis, and P450SPα from Sphingomonas paucimobilis, as members of the CYP152 family8,9,10,11,12, employ H2O2 instead of O2 as the oxidant as well as the electron donor to catalyze corresponding reactions.
Among these P450 peroxygenases, OleTJE fatty acid decarboxylase has drawn special attentions due to its potential application in biological production of α-alkenes as either biofuels or biomaterials. Catalytically, OleTJE mainly decarboxylates medium-to-long chain (C12-C20) fatty acids to generate terminal olefins (C11-C19) using H2O2 as cofactor. It also catalyzes α- and β-hydroxylation of fatty acids as side reactions8,13 (Fig. 1). This single-step transformation from free fatty acids to α-alkenes likely represents the most straightforward and efficient route for biosynthesis of aliphatic hydrocarbons, thus being an promising system to be engineered for cost-effective and environmentally sustainable production of fossil fuel alternatives in the future8,14,15.
Since its discovery by Rude et al. in 201113, intensive studies have been conducted in order to figure out the substrate specificity16,17,18, the reaction type selectivity (decarboxylation or hydroxylation)19, and the unique decarboxylation mechanism of OleTJE8,9,14,15,16. Recently, our laboratory uncovered the H2O2-independent activity of OleTJE. In addition to H2O2, OleTJE is also able to perform catalysis using the O2/redox partner/NADPH system. This result has important mechanistic implication and biotechnological significance8. Using alternative redox systems, this P450 enzyme showed different substrate specificity with the preferred substrate being C12, C14, or C18 fatty acids8. Makris and coworkers, using transient kinetic isotope effect analysis, recently reported that OleTJE catalysis is initiated by the formation of an iron(IV)-oxo π cation radical (Compound I)15. Fatty acid decarboxylation is likely resulted from the subsequent hydrogen abstraction from the Cβ position of substrate forming a stable Fe4+-OH species (compound II), which provides a rationale for the final carbon-carbon scission reaction15,20. Moreover, the crystal structure of OleTJE in complex with eicosanoic acid (C20) strongly suggested an essential role of the active site residues Arg245 and His85 for catalysis and reaction type selectivity, respectively9.
These studies have significantly advanced the understanding on the structural basis and catalytic mechanism of OleTJE. However, there remain a number of unsolved problems: What are the catalytic residues of OleTJE? What are the key amino acids determining whether decarboxylation or hydroxylation would occur? Is it possible to further improve the decarboxylation activity of OleTJE for practical application? Attempting to address these questions, in this work, we performed systematic mutagenesis analysis of four key residues including Arg245, Cys365, His85, and Ile170 to elucidate their functionality. Furthermore, a select group of redox partner proteins were screened in order to identify an optimal decarboxylation system.
Results and Discussion
Mutagenesis analysis of OleTJE
In the CYP152 family, an arginine residue has been found to be absolutely conserved (Supplementary Fig. S1). In the crystal structures of OleTJE, P450BSβ, and P450SPα9,10,21, the fixation of fatty acid substrate in active site all relies on the salt bridges between the guanidyl group of this arginine (Arg241 in P450SPα, Arg242 in P450BSβ, and Arg245 in OleTJE) and the terminal carboxyl group of fatty acids. Moreover, this arginine located near the heme iron reaction center is thought to be responsible for activating the iron(III)-bound H2O2 via acid-base catalysis to synthesise Compound I10.
Specifically in OleTJE, the guanidyl group of Arg245 was observed to be only 2.8 Å away from the carboxyl group of arachidic acid (Fig. 2B). To test if this residue is required for OleTJE activity, a series of single point mutants including R245A, R245Q, R245H, R245L, R245E and R245K were constructed. In the presence of 220 μM H2O2 and 200 μM myristic acid (C14) as substrate, the activities of all Arg245 mutants (2 μM) were compared in vitro using the wild type enzyme as positive control. As expected, except for R245K retaining marginal hydroxylation activity perhaps due to the similar chemical property of Lys and Arg, all other mutants were completely inactive (Table 1). These results clearly indicate an essential role of this arginine residue in OleTJE catalysis.
(A) Structural superimposition of OleTJE (in purple, PDB ID code 4L40) and P450BSβ (in grey, PDB ID code 1IZO); (B) Comparison of substrate binding pockets between OleTJE and P450BSβ. Red: heme iron; yellow: eicosanoic acid for OleTJE; green: palmitic acid for P450BSβ; purple: major active site residues in OleTJE; grey: major different amino acids in P450BSβ.
All P450 enzymes including CYP152 peroxygenases unanimously use a cysteine residue to coordinate with the heme iron. By contrast, almost all enzymes (except for chloroperoxidase) in non-P450 peroxygenase superfamily employ a histidine as the iron-coordinating ligand3,7. In consideration of the key role in O-O scission of the His ligand for peroxygenases22, we replaced Cys365 of OleTJE with His to investigate the impact on its peroxygenase activity. As a result, no activity was detected for the OleTJE C365H mutant (Table 1), which indicates this Cys ligand is critical for OleTJE and perhaps other P450 peroxygenases.
Among all CYP152 members that have been biochemically characterized so far, OleTJE is the only one that predominantly catalyzes fatty acid decarboxylation with α- and β-hydroxylation as side reactions. Other CYP152 enzymes including P450SPα12, P450BSβ23, and CYP-MP24 were identified as fatty acid hydroxylases primarily. P450SPα only hydroxylates fatty acids at α-position12. P450BSβ generates α-hydroxy and β-hydroxy fatty acids as major products, and a small amount of α-alkenes23. CYP-MP is able to introduce the hydroxyl group at α-, β-, γ-, δ-, and ε-position of C12-C18 fatty acids, while it only displayed minor decarboxylation activity against myristic acid (C14) and palmitic acid (C16)24. Taken together, it is important to identify the key amino acid residues, structural elements or any other factors responsible for the reaction type selectivity of P450 fatty acid decarboxylase/hydroxylase.
Comparatively, OleTJE and P450BSβ show 41%/60% amino acid sequence identity/similarity. Their overall structures and active site compositions are highly similar to each other (Fig. 2). The only two obviously different residues in their active sites are His85 (Gln85 in P450BSβ) and Ile170 (Val170 in P450BSβ), which reside at the two sides of the carboxyl group of substrate with a distance of 5.1 Å and 3.4 Å, respectively, in the crystal structure of OleTJE (Fig. 2). Thus, we hypothesize that these two amino acids might be related to the decarboxylation/hydroxylation selectivity.
To test this hypothesis, saturation mutagenesis of His85 and Ile170 in OleTJE was performed, and the in vitro activities of H85X and I170X variants towards myristic acid were evaluated (Fig. 3). As results, 11 out of 19 H85X variants were completely dead mutants. The rest 8 mutants unanimously lost their decarboxylation activities, while retaining varying hydroxylation activities (Fig. 3A). Notably, the two substitutions with an amide side chain (H85Q and H85N) retained most hydroxylation activity for unknown reasons.
Decarboxylation and hydroxylation reactions catalyzed by OleTJE and its mutants H85X (A) and I170X (B). Reaction conditions: wild type or mutant enzymes (2 μM), H2O2 (220 μM), and myristic acid (200 μM) in 200 μl desalting buffer were incubated at 30 °C for 16 h. All experiments were performed in duplicate.
Previously, Rude et al. proposed the importance of His85 for the decarboxylation activity of OleTJE based on the result that the Q85H mutation of P450BSβ enhanced its decarboxylation activity towards palmitic acid for about 50%13, which led to an increase in the ratio of decarcarboxylation to hydroxylation from 0.19 in wild type P450BSβ to 0.30 in the P450BSβ Q85H mutant. In this work, we confirmed the necessity of His85 for the decarboxylation activity of OleTJE. This is well consistent with the mechanism proposed by Belcher et al., in which His85 could act as a proton donor to compound I. This proton donation resulting in the protonated compound II was thought to be required for generation of the carboxylate radical for homolytic scission of the substrate C-Cα bond, thereby forming the terminal alkene. In the absence of proton donor, hydroxylation would be the only reaction9,20.
Similarly, none of the I170X mutants showed any decarboxylation activity against myristic acid. However, 11 out of 19 variants were able to catalyze the α- and/or β-hydroxylation to different extents (Fig. 3B). Thus, the mutagenesis analysis of His85 and Ile170 clearly indicates that these two residues adjacent to the carboxyl end of substrate are key factors for fatty acid decarboxylation.
Together with Arg245, these three amino acids are likely involved in the exact substrate positioning required for decarboxylation, explaining why conserved substitutions (e.g. I170V or I170L) also abolished the decarboxylation activity. In this regard, it might be highly challenging to rationally design a better version of OleTJE that is more selective for decarboxylation than hydroxylation without compromising the total activity, at least based on the current knowledge on the structure-function relationship of P450 fatty acid decarboxylases. A recent study of site-directed mutagenesis of OleTJE at selected sites lining the substrate binding pocket also proved difficulty in improving OleTJE activities towards structurally different aromatic carboxylic acid substrates. Only meager improvements (less than 1-fold) were observed in the few positively responded mutants (F79L and F294A)17. To overcome these rational design challenges, random gene mutagenesis or DNA shuffling coupled to high-throughput screening could be a more feasible strategy.
In vitro decarboxylation activity of OleTJE supported by different redox partners
When OleTJE was first identified to be a P450 fatty acid decarboxylase with potential application in the field of biofuels, it was thought to be an obligate peroxygenase as P450SPα and P450BSβ. However, our laboratory recently revealed the H2O2-independent activity of OleTJE (i.e. the activity depending on O2/redox partner(s)/NAD(P)H). This discovery has initiated the development of different olefin producing systems based on OleTJE and alternative redox partner protein(s). For instance, we have shown that the flavodoxin/flavodoxin reductase from E. coli and the RhFRED reductase from Rhodococcus sp. NCIMB 9784 are capable of supporting the OleTJE activity both in vitro and in vivo8. Dennig et al. employed putidaredoxin and putidaredoxin reductase from Pseudomonas putida to achieve the decarboxylation of short-chain fatty acids (C4-C9) into corresponding α-alkenes in vitro14 (Fig. 1). More importantly, by taking advantage of heterologous P450 redox partners, the engineered E. coli8 and Saccharomyces cerevisiae25 cells with OleTJE expression were able to produce 97.6 mg/L and 3.7 mg/L total α-alkenes, respectively.
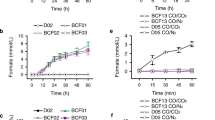
To identify the H2O2-independent activity of OleTJE with different redox partners, we in vitro screened a series of ferredoxins (Fdx) and ferredoxin reductases (FdR) derived from the cyanobacterial strain Synechococcus elongatus PCC 7942 and the Gram-positive bacterium Corynebacterium glutamicum ATCC 13032 (Supplementary DNA sequences of redox partners). Specifically, three FdRs (SeFdR-1 from S. elongates, and CgFdR-1 and CgFdR-2 from C. glutamicum) were individually coupled with ten Fdxs (SeFdx-1–7 from S. elongatus and CgFdx-1–3 from C. glutamicum), and 30 different combinations of redox partner proteins were mixed with OleTJE, myristic acid, and NADPH, respectively. The supportive activities of all redox partner combinations were compared to that of RhFRED and H2O2 (Fig. 4). Interestingly, all tested hybrid redox systems were able to support the in vitro decarboxylation activity of OleTJE to some extent, indicating the low selectivity of redox partners by this P450 fatty acid decarboxylase. The best combination turned out to be CgFdR-2 and SeFdx-6, which gave the highest conversion rate of 94.4% (Fig. 4). Using these two optimal redox partner proteins to mediate the electron transfer from NADPH, the steady-state kinetic parameters of OleTJE towards myristic acid were determined to be Km = 5.0 ± 2.4 μM, kcat = 2.2 ± 0.2 min−1, and kcat/Km = 0.4 μM−1 min−1 (Supplementary Fig. S2A). Comparatively, the values of Km and kcat were 24.2 ± 8.7 μM and 71.0 ± 8.4 min−1 (Supplementary Fig. S2B), respectively, when H2O2 was employed as the sole oxygen and electron donor. The kcat/Km value of 2.9 μM−1 min−1 greater than 0.4 μM−1 min−1 seemed inconsistent with the qualitative results that the CgFdR-2/SeFdx-6/NADPH redox system showed higher fatty acid to α-alkene conversion rate (94.4%) than that supported by H2O2 (49.5%). We reason this contradiction might be due to inactivation of OleTJE by H2O2 during prolonged incubation. Taken together, these results demonstrated efficient monooxygenase-like property of OleTJE to use the O2/redox partner(s)/NAD(P)H system, which is critical for the future investigation of the unique mechanism and better application of this enzyme.
Conclusion
We have systematically investigated the functions of three active site residues of OleTJE including Arg245, His85, and Ile170 by site-directed mutagenesis. It was found that they are all required for the decarboxylation activity of OleTJE, presumably by forming a salt-bridge with the substrate carboxyl group (Arg245), by acting as a proton donor (His85), and by precisely coordinating substrate positioning in the active site (Ile170). We also studied the H2O2-independent activity of OleTJE and revealed a series of heterologous redox partners capable of supporting its decarboxylation activity efficiently in vitro. These results not only further our understanding on the unique decarboxylative mechanism of OleTJE, but also serve as a guide for further bioengineering of this P450 system and the future industrial application.
Methods
Reagents
The recombinant plasmid pET28b-oleTJE for overexpression of the P450 enzyme OleTJE was constructed by our laboratory previously8. Fatty acids (myristic acid and heptadecanoic acid), 1-tridecene authentic standards, and derivatizing reagent BSTFA-TMCS were purchased from TCI (Shanghai, China). Antibiotics and isopropyl β-D-1-thiogalactopyranoside (IPTG) were obtained from Solarbio (Beijing, China). All restricted enzymes were purchased from Thermo Scientific (Shanghai, China). PrimeSTAR GXL DNA polymerase was obtained from Takara (Otsu, Japan). Kits used for DNA manipulation were bought from OMEGA Bio-Tek (Jinan, China) or Promega (Madison, WI, USA). Ni-NTA resin from Qiagen (Valencia, CA, USA), Millipore Amicon Ultra centrifugal fliters (Billerica, MA, USA) and PD-10 desalting columns purchased from GE Healthcare (Piscataway, NJ, USA) were used for protein purification.
Strains, plasmids and media
Escherichia coli DH5α cells were used for plasmid transformation and mutant screening. Escherichia coli BL21(DE3) was used for protein overexpression. The plasmid pET28b was used for gene cloning. E. coli cells were grown in Terrific Broth medium composed of 1.2% tryptone, 0.5% glycerol, 2.4% yeast extract, 0.23% KH2PO4 and 1.25% K2HPO4, supplemented with the selective antibiotic (50 μg/mL kanamycin), thiamine (1 mM) and rare salt solution for protein expression26.
Molecular cloning
With pET28b-oleTJE as template, the oleTJE mutants were constructed using the Quikchange mutagenesis method and cloned into pET28b vector. Mutagenesis primers are listed in Supplementary Table S1.
The coding DNA sequences of ferredoxin reductase CgFdR-1 and CgFdR-2 were amplified from Corynebacterium glutamicum ATCC 13032. The coding DNA sequences of seven ferredoxins SeFdx-1, SeFdx-2, SeFdx-3, SeFdx-4, SeFdx-5, SeFdx-6 and SeFdx-7 from Synechococcus elongatus PCC 7942 were codon-optimized by Genewiz (Suzhou, China) and cloned into pET28b. The other three ferredoxin genes encoding CgFdx-1, CgFdx-2 and CgFdx-3 were PCR amplified from Corynebacterium glutamicum ATCC 13032. The primers are listed in Supplementary Table S2. The NdeI and XhoI restriction sites were used for cloning into the NdeI/XhoI pre-treated pET28b to obtain corresponding expression vectors.
Overexpression and purification of proteins
The recombinant expression plasmids were transformed into E. coli BL21(DE3). After cultivation in LB medium containing 50 μg/mL kanamycin at 37 °C, 220 rpm overnight, 1% volume of the seed culture was used to inoculate 1 L Teffific Broth medium containing 50 μg/mL kanamycin, 1 mM thiamine and rare salt solution. When the OD600 reached 0.6–0.8, IPTG was added to a final concentration of 0.2 mM for induction, followed by shaking incubation at 18 °C for 20 h.
After harvesting by centrifugation at 4 °C, 6000 rpm for 10 min, cells were stored at −80 °C for 30 min. Then cells were thawed and re-suspended in 40 mL lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10% glycerol and 10 mM imidazole, pH 8.0) and sonicated on a JY92-IIDN ultra-sonicator for 30 min with a 5 s on and 5 s off pulse. The whole cell lysates were centrifuged at 12,000 rpm for 30 min at 4 °C. The clarified cell lysates were collected and incubated with 1 mL Ni-NTA resin under gentle shaking at 4 °C for 1–2 h. The mixture was then loaded onto an empty column and washed with approximately 100 mL wash buffer (50 mM NaH2PO4, 300 mM NaCl, 10% glycerol and 20 mM imidazole, pH 8.0) until no proteins were detected in the flow-through. The bound protein was eluted by 10 mL elution buffer (50 mM NaH2PO4, 300 mM NaCl, 10% glycerol and 250 mM imidazole, pH 8.0) and then concentrated using a Millipore Ultra-filter (30 K). The concentrated protein solution was loaded onto a PD-10 column, which was used for removal of imidazole and buffer exchange into desalting buffer (50 mM NaH2PO4 and 10% glycerol, pH 7.4). The eluent was flash-frozen by liquid nitrogen and stored at −80 °C for later use.
Determination of enzyme concentration
The ferric-CO complex of P450 enzyme was prepared by slow bubbling of carbon monoxide gas into a solution of purified ferric P450 for 1 min. Then its UV-visible absorption spectrum from 300 nm to 600 nm was recorded on a UV-visible spectrophotometer DU800 (Beckman Coulter, Fullerton, CA, USA). Following reduction by sodium dithionite, the corresponding spectrum of reduced ferrous-CO adducts was recorded. The functional concentration of P450 was calculated from the CO-bound reduced differential spectrum using a molar extinction coefficient (ε450–490) of 91 mM−1.cm−1 27.
The concentration of purified ferredoxin and ferredoxin reductase was determined by monitoring the absorbance at 325 nm and 454 nm, respectively, and using their corresponding molar extinction coefficient 15.4 mM−1.cm−1 (ε325) and 11.3 mM−1.cm−1 (ε454)28.
In vitro enzymatic assays of OleTJE variants with H2O2 as sole cofactor
Fatty acid decarboxylation and hydroxylation assays of purified OleTJE variants were performed in 200 μL reaction mixture containing 2 μM OleTJE or OleTJE mutants, 220 μM H2O2, and 200 μM myristic acid as substrate in desalting buffer. The reactions were carried out at 30 °C for 16 h and then quenched with 20 μL of 10 M HCl. After adding 500 μM heptadecanoic acid as the internal standard, 150 μL of ethyl acetate was used for extraction. The mixture was separated into aqueous and organic phase by centrifugation at 14,000 rpm for 10 min. 70 μL of the organic phase was derivatized with an equal volume of N,O-bis (trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane (BSTFA-TMCS) at 70 °C for 15 min. Samples were then analyzed using gas chromatography-mass spectroscopy (GC-MS).
In vitro enzymatic assays of OleTJE with the O2/NAD(P)H/redox partners system
Fatty acids decarboxylation of purified OleTJE supported by various heterologous redox partners were performed in 100 μL desalting buffer containing 1 μM OleTJE, 5 mM NAD(P)H, 400 μM myristic acid substrate, 20 μM ferredoxin and 20 μM ferredoxin reductase. The reactions were carried out at 30 °C for 16 h and then quenched with 10 μL of 10 M HCl. After adding 500 μM heptadecanoic acid as the internal standard, 150 μL of ethyl acetate was used for extraction. Samples were then analyzed using GC-MS.
Steady-state kinetics
For the reaction system using H2O2 as the sole oxygen and electron donor, the standard reactions containing 50 nM of OleTJE, 20–100 μM myristic acid in 1.2 mL of desalting buffer (50 mM NaH2PO4 and 10% glycerol, pH 7.4) were pre-incubated at 30 °C for 5 min. The reactions were then initiated by adding 220 μM H2O2. For the O2/CgFdR-2/SeFdx-6/NADPH system, the standard reactions containing 100 nM of OleTJE, 10–50 μM myristic acid, 2 μM CgFdR-2 from C. glutamicum ATCC 13032 and 2 μM SeFdx-6 from Synechococcus elongatus PCC 7942 in 1.2 mL of desalting buffer (50 mM NaH2PO4 and 10% glycerol, pH 7.4) were pre-incubated at 30 °C for 5 min. The reactions were then initiated by adding 500 μM NADPH. For all reactions, 200 μL aliquots were taken at four different time points to be stopped by adding 1/10 volume of 10 M HCl. Sample extraction was performed as above with 500 μM heptadecanoic acid as the internal standard. For Km and Vmax determinations, the initial velocity of product formation was monitored by GC-MS. Kinetic data from duplicated experiments were fit into Michaelis-Menten equation using Origin 8.0 program.
Analytical method
Qualitative and quantitative analysis of the products were performed by GC-MS29. To detect α-alkenes, an Agilent 7890A gas chromatography equipped with HP-INNOWAX (Agilent Technologies, Inc., cross-linked polyethylene glycerol, Santa Clara, CA, USA; 0.25 µm film thickness, 30 m by 0.25 mm) column was adopted. The column heating program was as follows: the initial temperature of oven was set to 40 °C for 4 min, then increased at a rate of 10 °C/min to 250 °C, and hold for 15 min. The α- and β-hydroxy fatty acids detection was carried out using the Agilent J&W DB-5 MS column (0.25 μm film thickness, 30 m by 0.25 mm). Furthermore, an Agilent 5975C MSD quadrupole mass spectrometer with a scan range from 50 to 500 m/z under electron ionization condition (70 eV) was coupled to the GC. The oven temperature was 50 °C initially and ramped up to 300 °C at the above mentioned rate, then 300 °C for 5 min. Quantification was performed using the corresponding authentic standard compounds and heptadecanoic acid as the internal standard.
Additional Information
How to cite this article: Fang, B. et al. Mutagenesis and redox partners analysis of the P450 fatty acid decarboxylase OleTJE. Sci. Rep. 7, 44258; doi: 10.1038/srep44258 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Denisov, I. G., Makris, T. M., Sligar, S. G. & Schlichting, I. Structure and chemistry of cytochrome P450. Chem. Rev. 105, 2253–2278 (2005).
Hrycay, E. G. & Bandiera, S. M. The monooxygenase, peroxidase, and peroxygenase properties of cytochrome P450. Arch. Biochem. Biophys. 522, 71–89 (2012).
Sono, M., Roach, M. P., Coulter, E. D. & Dawson, J. H. Heme-Containing Oxygenases. Chem. Rev. 96, 2841–2888 (1996).
Guengerich, F. P. & Munro, A. W. Unusual Cytochrome P450 Enzymes and Reactions. J. Biol. Chem. 288, 17065–17073 (2013).
Domanski, T. L. & Halpert, J. R. Analysis of Mammalian Cytochrome P450 Structure and Function by Site-Directed Mutagenesis. Curr. Drug Metab. 2, 117–137 (2001).
Furge, L. L. & Guengerich, F. P. Cytochrome P450 enzymes in drug metabolism and chemical toxicology: An introduction. Biochem. Mol. Biol. Educ. 34, 66–74 (2006).
Sundaramoorthy, M., Terner, J. & Poulos, T. L. The crystal structure of chloroperoxidase: A heme peroxidase-cytochrome P450 functional hybrid. Structure 3, 1367–1377 (1995).
Liu, Y. et al. Hydrogen peroxide-independent production of alpha-alkenes by OleT(JE) P450 fatty acid decarboxylase. Biotechnol. Biofuels 7, 28–39 (2014).
Belcher, J. et al. Structure and biochemical properties of the alkene producing cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 bacterium. The J. Biol. Chem. 289, 6535–6550 (2014).
Fujishiro, T. et al. Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position. J. Biol. Chem. 286, 29941–29950 (2011).
Shoji, O. et al. Hydrogen peroxide dependent monooxygenations by tricking the substrate recognition of cytochrome P450BSbeta. Angew. Chem. Int. Ed. 46, 3656–3659 (2007).
Matsunaga, I., Sumimoto, T., Ueda, A., Kusunose, E. & Ichihara, K. Fatty acid-specific, regiospecific, and stereospecific hydroxylation by cytochrome P450 (CYP152B1) from Sphingomonas paucimobilis: Substrate structure required for alpha-hydroxylation. Lipids 35, 365–371 (2000).
Rude, M. A. et al. Terminal olefin (1-alkene) biosynthesis by a novel p450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 77, 1718–1727 (2011).
Dennig, A. et al. Oxidative Decarboxylation of Short-Chain Fatty Acids to 1-Alkenes. Angew. Chem. Int. Ed. 54, 8819–8822 (2015).
Grant, J. L., Hsieh, C. H. & Makris, T. M. Decarboxylation of fatty acids to terminal alkenes by cytochrome P450 compound I. J. Am. Chem. Soc. 137, 4940–4943 (2015).
Hsieh, C. H. & Makris, T. M. Expanding the substrate scope and reactivity of cytochrome P450 OleT. Biochem. Biophys. Res. Commun. 476, 462–466 (2016).
Wang, J. B., Lonsdale, R. & Reetz, M. T. Exploring substrate scope and stereoselectivity of P450 peroxygenase OleT(JE) in olefin-forming oxidative decarboxylation. Chem. Commun. 52, 8131–8133 (2016).
Dennig, A. et al. Enzymatic Oxidative Tandem Decarboxylation of Dioic Acids to Terminal Dienes. European J. Org. Chem. 21, 3473–3477 (2016).
Faponle, A. S., Quesne, M. G. & de Visser, S. P. Origin of the Regioselective Fatty-Acid Hydroxylation versus Decarboxylation by a Cytochrome P450 Peroxygenase: What Drives the Reaction to Biofuel Production? Chem-Eur. J. 22, 5478–5483 (2016).
Grant, J. L., Mitchell, M. E. & Makris, T. M. Catalytic strategy for carbon-carbon bond scission by the cytochrome P450 OleT. Proc. Natl. Acad. Sci. USA 113, 10049–10054 (2016).
Lee, D. S. et al. Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis. Crystallographic, spectroscopic, and mutational studies. J. Biomol. Screen. 278, 9761–9767 (2003).
Yi, X. W., Mroczko, M., Manoj, K. M., Wang, X. T. & Hager, L. P. Replacement of the proximal heme thiolate ligand in chloroperoxidase with a histidine residue. Proc. Natl. Acad. Sci. USA 96, 12412–12417 (1999).
Matsunaga, I., Ueda, A., Fujiwara, N., Sumimoto, T. & Ichihara, K. Characterization of the ybdT gene product of Bacillus subtilis: Novel fatty acid beta-hydroxylating cytochrome P450. Lipids 34, 841–846 (1999).
Amaya, J. A., Rutland, C. D. & Makris, T. M. Mixed regiospecificity compromises alkene synthesis by a cytochrome P450 peroxygenase from Methylobacterium populi . J. Inorg. Biochem. 158, 11–16 (2016).
Chen, B., Lee, D. Y. & Chang, M. W. Combinatorial metabolic engineering of Saccharomyces cerevisiae for terminal alkene production. Metab. Eng. 31, 53–61 (2015).
Anzai, Y. et al. Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics. Chem. Biol. 15, 950–959 (2008).
Johnston, W. A., Huang, W. L., De Voss, J. J., Hayes, M. A. & Gillam, E. M. J. Quantitative whole-cell cytochrome P450 measurement suitable for high-throughput application. J. Biomol. Screen. 13, 135–141 (2008).
Chun, Y.-J. et al. Electron Transport Pathway for a Streptomyces Cytochrome P450: cytochrome P450 105D5-catalyzed fatty acid hydroxylation in Streptomyces coelicolor A3(2). J. Biol. Chem. 282, 17486–17500 (2007).
Guan, W. et al. Quantitative analysis of fatty-acid-based biofuels produced by wild-type and genetically engineered cyanobacteria by gas chromatography-mass spectrometry. J. Chromatogr. A 1218, 8289–8293 (2011).
Acknowledgements
This work was supported by the National Science Foundation of China (NSFC 31422002, 31270855 and 21406250), Shandong Provincial Natural Science Foundation Grant JQ201407, and Applied Basic Research Programs of Science and Technology of Qingdao (15-9-1-106-jch). We are grateful to Prof. Xuefeng Lu at the Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences for kindly providing the genomic DNA of Synechococcus elongatus PCC 7942.
Author information
Authors and Affiliations
Contributions
S.L. and H.X. conceived of the study. B.F., H.X., Y.L., H.C., C.W. and S.L. designed the experiments. B.F., H.X., Y.L., F.Q., W.Z., H.C., Y.W., and W.Y. performed the experiments including plasmid construction, protein overexpression, purification, mutagenesis, product characterization and enzymatic assays. B.F., Y.L. and C.W. carried out GC-MS analysis. B.F., H.X. and S.L. drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Fang, B., Xu, H., Liu, Y. et al. Mutagenesis and redox partners analysis of the P450 fatty acid decarboxylase OleTJE. Sci Rep 7, 44258 (2017). https://doi.org/10.1038/srep44258
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44258
This article is cited by
-
Establishing an enzyme cascade for one-pot production of α-olefins from low-cost triglycerides and oils without exogenous H2O2 addition
Biotechnology for Biofuels (2020)
-
Biochemical characterization of three new α-olefin-producing P450 fatty acid decarboxylases with a halophilic property
Biotechnology for Biofuels (2019)
-
Different Behaviors of a Substrate in P450 Decarboxylase and Hydroxylase Reveal Reactivity-Enabling Actors
Scientific Reports (2018)
-
In vitro oxidative decarboxylation of free fatty acids to terminal alkenes by two new P450 peroxygenases
Biotechnology for Biofuels (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.