Abstract
Opitz trigonocephaly C syndrome (OTCS) is a rare genetic disorder characterized by craniofacial anomalies, variable intellectual and psychomotor disability, and variable cardiac defects with a high mortality rate. Different patterns of inheritance and genetic heterogeneity are known in this syndrome. Whole exome and genome sequencing of a 19-year-old girl (P7), initially diagnosed with OTCS, revealed a de novo nonsense mutation, p.Q638*, in the MAGEL2 gene. MAGEL2 is an imprinted, maternally silenced, gene located at 15q11-13, within the Prader-Willi region. Patient P7 carried the mutation in the paternal chromosome. Recently, mutations in MAGEL2 have been described in Schaaf-Yang syndrome (SHFYNG) and in severe arthrogryposis. Patient P7 bears resemblances with SHFYNG cases but has other findings not described in this syndrome and common in OTCS. We sequenced MAGEL2 in nine additional OTCS patients and no mutations were found. This study provides the first clear molecular genetic basis for an OTCS case, indicates that there is overlap between OTCS and SHFYNG syndromes, and confirms that OTCS is genetically heterogeneous. Genes encoding MAGEL2 partners, either in the retrograde transport or in the ubiquitination-deubiquitination complexes, are promising candidates as OTCS disease-causing genes.
Similar content being viewed by others
Introduction
Opitz C syndrome (or Opitz-trigonocephaly, OTCS; MIM #211750) is a rare, severe and heterogeneous disorder with some 60 cases described worldwide1. Manifestations in this syndrome affect all body systems, are highly variable among different patients, and comprise psychomotor delay, trigonocephaly and a characteristic combination of facial dysmorphisms including retrognathia, upslanted palpebral fissures, and epicanthic folds. Short neck, joint contractures, cryptorchidism, congenital heart malformations and pancreatic and renal involvement may also be present. Commonly, OTCS patients are hypotonic, have arthrogryposis and seizures2. The syndrome has a high infant mortality rate and around 50% of patients die during the first year of life mostly due to respiratory failure and cardiovascular malformations2. OTCS is similar to other syndromes, mainly to Bohring-Opitz syndrome (BOS or C-like syndrome; MIM #605039), a more severe disorder3. The molecular bases of OTCS are still unknown (although CD96 has been proposed as an OTCS gene3, its implication has been recently questioned)4,5. Likewise, the pattern of inheritance is unclear, with many sporadic cases suggestive of a dominant inheritance and some cases that point to autosomal recessive inheritance because of affected sibs or parental consanguinity. Genetic heterogeneity and clinical variability should be considered in OTCS as in other developmental delay and intellectual disability syndromes.
We previously performed whole exome sequencing (WES) in 5 OTCS patients with normal CD96 and ASXL1 genes5. Here we present the results of one of them, a 19 year-old Spanish woman, in whom we found a de novo nonsense mutation in MAGEL2. Whole genome sequencing (WGS), performed in parallel, gave a concordant result after analyzing the exonic data. The MAGEL2 gene maps to the imprinted Prader-Willi Critical Region (PWCR) on chromosome 15. Several de novo truncating mutations, similar to the one described here, have been reported previously in patients with Schaaf-Yang syndrome (SHFYNG, MIM #615547)6,7, a condition with some resemblances to Prader-Willi syndrome, and in two patients with a distinct severe arthrogryposis phenotype8. Very recently, Fountain et al.9 described 18 additional SHFYNG patients with similar mutations.
Here we present a detailed phenotypic description of this female patient, originally diagnosed as OTCS, and we describe her phenotype in light of the main features of SHFYNG.
Results
Clinical Report
The patient, a 19-year-old woman, is the first and only daughter of a non-consanguineous couple. She was born at full-term by caesarean section with a birth weight of 2.6 kg (34th centile), length of 48 cm (49th centile) and head circumference of 34 cm (61st centile). She had neonatal respiratory depression (Apgar scores 1/5), hypotonia, contractures of fingers and toes and, less evident, bilateral clubfoot. At 1 month she had a cardiopulmonary arrest and bradycardia. In the following months trigonocephaly due to premature metopic suture fusion was noted (Fig. 1A) and at age 2y she was treated neurosurgically. She has had feeding difficulties and oropharyngeal dysphagia and currently eats soft food and liquids, but nothing solid.
There was severe delay in achieving motor milestones and she started to walk with support around age 11y. She has severe intellectual disability with no language, as well as constant sleep disturbances such as long periods of insomnia (up to 72 hours) and difficulties initiating or maintaining sleep. Temperature instability, profuse sweating and excessive salivation occur frequently. Age at menarche was 14y. At 18y she developed sleep apnea and episodic hyperventilation similar to the pattern described in Pitt-Hopkins syndrome. She has suffered multiple infections during her life.
At present (age 19y) her stature, weight and head circumference are below the 3rd centile, and she shows frontal cowlick, strabismus, short nose, slightly anteverted nares, macrostomia, thick palatal and alveolar ridges, teeth malposition (Fig. 1B and C), wide-spaced nipples, hypoplasia of labia majora with prominent clitoris, mild limitation of elbow extension, hands with abnormal palmar creases, hand and feet camptodactyly (Fig. 1D–H) and mild webbing, asymmetric thorax and marked lordosis.
The patient was tentatively diagnosed at 2 years of age as affected with OTCS, confirmed by one of us (JMO) at an Opitz C Syndrome Parent Support Group and Scientific meeting, held in Chicago, Illinois, USA, in 1998.
Complementary Analyses
Complementary tests and their results (in parentheses) are as follows: karyotype (46, XX); metabolic screening (normal); echocardiogram (normal); cranial CT scan (2y: trigonocephaly); gynaecologic ultrasound (8y: infantile uterus and normal ovaries) cerebral MRI (10y and 13y: thin corpus callosum, inferior vermis hypoplasia, mild brachycephaly, hypophysis of normal size); methylation study of the SNRPN locus within the Prader-Willi region by M-PCR (normal); FISH of the centromeric region of chromosome 12 (normal, Pallister-Killian Syndrome ruled out); sequencing of TCF4 (no pathogenic mutations, ruling out Pitt-Hopkins syndrome); SNP array (250,000 SNPs, normal).
Exome and Genome Sequence Analysis
On average, the mean WES coverage for the P7 trio was of 59.1, and 93.2% of the target region was covered by at least 10 reads (C10). See Table S1 for further details.
The main WES result was a de novo heterozygous mutation in the MAGEL2 gene, among some variants of unknown significance in other genes (reported in Table S2). This result was confirmed by an independent WGS.
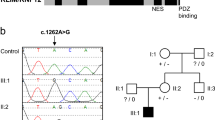
The MAGEL2 mutation consists of a c.1912C > T transition, which leads to the substitution of a glutamine (Q) residue by a STOP codon (p.Q638*) (Fig. 2). Since MAGEL2 is imprinted and maternally silenced, we experimentally confirmed that the change was present on the paternal chromosome.
WH-A and B: Winged-Helix A and B (residues 1013-1097 & 1098-1184); Ext: Winged-Helix B Extension (residues 1185-1227); USP7 BS (USP7 Binding Site, residues 949-1004). In italics, mutation p.Q638* found in the OTCS patient described here and in a SHFYNG patient9. Bold and grey represent mutations found in severe arthrogryposis patients8,9.
MAGEL2 Analysis in a Cohort of 9 OTCS Patients
The MAGEL2 coding region plus 340 bp of the 5′-UTR and 142 bp of the 3′-UTR were Sanger sequenced in 9 other patients diagnosed as OTCS. Patient P2 bore the missense mutation p.A632T [c.1894G > A; ExAC MAF: 0.00003337 (1/29966 individuals)] and patient P12a presented a polymorphic 21-bp in-frame deletion (rs774629250, MAF: 0.0002). Both mutations were present in a heterozygous state and inherited from the respective mothers and, thus, are in the allele predicted to be silenced. We did not find any putatively pathologic mutation in these 9 patients.
Discussion
In our study, by means of whole-exome and genome sequencing, we have found a de novo truncating mutation in the maternally-silenced MAGEL2 gene in a patient clinically diagnosed as Opitz C. The mutation occurred on the paternal chromosome. No other MAGEL2 pathogenic mutations were found in any of the remaining 9 OTCS patients that we were able to test. Given the uncertain role of CD96 in OTCS, as mentioned above, MAGEL2 might be the first gene clearly associated with Opitz C syndrome. The fact that we did not find any other MAGEL2 mutation in 9 additional OTCS patients is an indication of genetic heterogeneity of this syndrome.
Truncating mutations in the MAGEL2 gene were identified previously in two distinct conditions, the Schaaf-Yang syndrome, a relatively mild condition with some resemblance to the Prader-Willi syndrome (see ref. 10 for further comparisons), and in patients with a severe form of arthrogryposis with reduced fetal movement and perinatal death8. Recently, Fountain et al.9, described a large series of SHFYNG patients, all with a truncating MAGEL2 mutation on the paternal chromosome. Interestingly, they reported three familial cases in which the mutation was inherited from the father. The cosegregation of the mutations and phenotypes in these pedigrees further confirm the pathogenicity of MAGEL2 truncation.
The patient described here (P7) bears a de novo c.1912C > T (p.Q638*) mutation, identical to the mutation present in patient 4 of Fountain et al.9. In fact, our patient P7 bears good phenotypic resemblance with SHFYNG. She presented with developmental delay, which is observed among all surviving patients with MAGEL2 mutations, neonatal hypotonia, feeding problems in infancy and arthrogryposis or joint contractures, all traits common to the majority of the patients with MAGEL2 mutations. Autism spectrum disorder (ASD) -common in MAGEL2-mutated patients- could not be evaluated due to her severe intellectual disability. In spite of the age difference, our patient and patient 4 of Fountain et al.9, with the same MAGEL2 mutation, share developmental and intellectual disability, feeding problems, neonatal hypotonia, contractures, minor facial anomalies, small hands, sleep apnea and temperature instability. Patient P7 is similar in age and sex to patient 18 of Fountain et al.9. They are concordant for several features including female hypogenitalism, but discordant with respect to behaviour, sleep apnea, temperature instability and seizures. Worthy of note are a few features present in our patient and not described in the other patients with MAGEL2 mutations. These include insomnia and sleep difficulties, also observed in Magel2 null mice10, thick palatal and alveolar ridges, weak cry, tooth malposition, trigonocephaly, excessive salivation, episodic hyperventilation and multiple dislocations, many of which are characteristic of OTCS patients.
Despite the high clinical variability observed among patients with mutations in MAGEL2, all bear truncating mutations that lead to the formation of a shorter version of the protein lacking the MAGE homology domain (MHD) (Fig. 2). So far, a total of 12 different mutations has been reported. Notably, they cluster in the middle and 3′ portion of the gene, between codons 541 and 1079, of the 1249 of the full MAGEL2 coding region. There are some recurrent mutations, suggesting the existence of hotspots. It is interesting that one of these hotspots gives rise to two different mutations (c.1996dupC and c.1996delC). One was reported in 12 patients from 8 unrelated families and another in 4 patients from two families. The phenotype associated with the latter is a severe neonatally lethal arthrogryposis, while the former was never associated with this phenotype. This spectrum of phenotypes, with different natural histories, might be gathered under the genetic umbrella of MAGELopathies, in a similar way to RASopathies11.
MAGEL2 belongs to the MAGE (melanoma antigen) domain containing family of proteins12, and is known to bind and to enhance the activity of the TRIM27 E3 RING ubiquitin ligase. The MAGEL2-TRIM27 complex, which includes and is regulated by USP7, appears to play a key role in the retrograde transport from the endosome to the trans-Golgi network, an important cellular process which facilitates the recycling of a variety of proteins. Other critical components of this retrograde transport are the three members of the retromer complex (VPS26, VPS29, and VPS35) and the WASH regulatory complex (SHRC), which consists of at least five core factors: CCDC53, FAM21, SWIP, Strumpellin, and WASH13,14. Interestingly, mutations in different members of the retrograde transport lead to diseases characterized by intellectual disability and developmental delay (Table 1). Mutations in the USP7 are associated with a syndromic form of intellectual disability including features of ASD, hypotonia and seizures15. In addition, mutations in SWIP (KIAA1033) and strumpellin (KIAA0196) were found associated with intellectual disability. In particular, mutations in SWIP have been identified as responsible for a nonsyndromic, autosomal recessive form of intellectual disability with short stature in one family16, while strumpellin was associated with two different diseases. On one hand, mutations that lead to the loss of the C-term of the protein were associated with the Ritscher-Schinzel syndrome (RTSC1, MIM#220210), an autosomal recessive condition characterized by intellectual disability, craniofacial abnormalities, congenital heart defects and cerebellar brain malformations17. On the other hand, missense mutations were associated with spastic paraplegia 8. In addition, other partners of the WASH complex have been associated with Parkinson disease (PD)18, Alzheimer disease19 and ASD20.
When looking at the photographs of SHFYNG patients9 and those of OTCS patients5, there are some obvious resemblances. The clinical overlap between these two syndromes suggests that some of the patients initially diagnosed as OTCS may be, in fact, SHFYNG patients and thus, MAGEL2 should be assessed in OTCS patients. We found only one out of ten OTCS patients with a truncating mutation in MAGEL2, but it is likely that other OTCS patients might bear mutations in this gene and we strongly recommend that it should be sequenced in all of them. MAGEL2 was not well captured in many of the exome assays and, in particular, in capture kits based on the 37 build of the genome the 5′ part of the gene is lacking. For example, Soden et al.7, investigating neurodevelopmental disorders, mentioned that they found mutations by WGS that were not observed in exome analyses. On the other hand, for MAGEL2-negative OTCS patients, the causal genes could be among those coding for other members of the retrograde transport complex or, more specifically, of the ubiquitination and deubiquitination complex. TRIM27 and USP7 form one such ubiquitination-deubiquitination complex21. In this regard, USP7 has been recently shown to regulate ASXL122, whose mutations are the cause of 50–70% of the cases of Bohring-Opitz syndrome5,23,24,25. These functional clues will surely help pinpoint the remaining OTCS causal genes in ongoing whole exome and genome sequencing studies.
Materials and Methods
Patients and Samples
The Spanish patient with the MAGEL2 mutation is P7 in Urreizti et al.7, together with the four other patients subjected to WES (P1, P2, P6 and P8) and two additional patients (P10a and P11). Three additional patients from three independent families (P12a, P13 and P14) were recruited from Malta, Australia and Spain, respectively, and diagnosed by one of us (either JMO or GN). Written informed consent was obtained from all patients, including a specific informed consent to publish images for patient P7. All methods were carried out in accordance with relevant guidelines and regulations and the Bioethics Committee of the University of Barcelona approved the protocols.
Exome Sequencing and Filtering
Genomic DNA from patient P7 and her parents was sequenced in the National Center of Genomic Analysis (CNAG; Barcelona, Spain) using the Illumina HiSeq-2000 platform. Exome capture was performed with Nimblegen SeqCap 64 Mb v3 (Roche; Mannheim; Germany). The samples where sequenced at a coverage of 50x. The data were analysed as described elsewhere26 (see Supplementary Methods).
Validation by PCR Amplification and Sanger Sequencing
A total of 75 variants was selected for validation. A primer pair for each fragment containing the position of the putative change was designed using Primer 327 (primer sequences and PCR conditions are available on demand). PCR reactions, purification and sequencing were performed as previously described5.
Genome Sequencing
The whole genomic DNA from the patient and her parents has been sequenced at the CRG Genomics unit. Libraries were prepared using the NEBNext Ultra DNA libray prep kit for Illumina. Sequencing was performed on an Illumina HiSeq2500 instrument, using paired-end 125 bp reads, at the coverage of 36–42x (see Supplementary Methods).
Phasing the de novo Mutation in MAGEL2
To assess the phase of the mutation a methylation-sensitive digestion was performed, as in Schaaf et al.6 with some modifications (see Supplementary Methods).
PCR Amplification and Sanger Sequencing of the MAGEL2 Gene
A region of 4.2 kb including the MAGEL2 coding region, 304 bp of the 5′-UTR and 143 of the 3′-UTR (GRCh38 ENSG00000254585) was amplified in 9 overlapping fragments prior to Sanger sequencing. Primer pairs and PCR conditions are summarized in Table S3.
Additional Information
How to cite this article: Urreizti, R. et al. A De Novo Nonsense Mutation in MAGEL2 in a Patient Initially Diagnosed as Opitz-C: Similarities Between Schaaf-Yang and Opitz-C Syndromes. Sci. Rep. 7, 44138;doi: 10.1038/srep44138 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Travan, L. et al. Opitz trigonocephaly syndrome presenting with sudden unexplained death in the operating room: a case report. J. Med. Case Rep. 5, 222 (2011).
Opitz, J. M. et al. Mortality and pathological findings in C (Opitz trigonocephaly) syndrome. Fetal Pediatr. Pathol. 25, 211–231 (2006).
Kaname, T. et al. Mutations in CD96, a member of the immunoglobulin superfamily, cause a form of the C (Opitz trigonocephaly) syndrome. Am. J. Hum. Genet. 81, 835–841 (2007).
Darlow, J. M., McKay, L., Dobson, M. G., Barton, D. E. & Winship, I. On the origins of renal cell carcinoma, vesicoureteric reflux and C (Opitz trigonocephaly) syndrome: A complex puzzle revealed by the sequencing of an inherited t(2;3) translocation. Eur. J. Hum. Genet. 21, 145 (2013).
Urreizti, R. et al. Screening of CD96 and ASXL1 in 11 patients with Opitz C or Bohring-Opitz syndromes. Am. J. Med. Genet. A 170, 24–31 (2016).
Schaaf, C. P. et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat. Genet. 45, 1405–1408 (2013).
Soden, S. E. et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 6, 265ra168 (2014).
Mejlachowicz, D. et al. Truncating Mutations of MAGEL2, a Gene within the Prader-Willi Locus, Are Responsible for Severe Arthrogryposis. Am140. J. Hum. Genet. 97, 616–620 (2015).
Fountain, M. D. et al. The phenotypic spectrum of Schaaf-Yang syndrome: 18 new affected individuals from 14 families. Genet. Med. 19, 45–52 (2017).
Devos, J., Weselake, S. V. & Wevrick, R. Magel2, a Prader-Willi syndrome candidate gene, modulates the activities of circadian rhythm proteins in cultured cells. J. Circadian Rhythms 9, 12 (2011).
Tidyman, W. E. & Rauen, K. A. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 19, 230–236 (2009).
Chomez, P. et al. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 61, 5544–5551 (2001).
Seaman, M. N. The retromer complex - endosomal protein recycling and beyond. J. Cell Sci. 125, 4693–4702 (2012).
Hao, Y. H. et al. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 152, 1051–1064 (2013).
Hao, Y. H. et al. USP7 Acts as a Molecular Rheostat to Promote WASH-Dependent Endosomal Protein Recycling and Is Mutated in a Human Neurodevelopmental Disorder. Mol. Cell 59, 956–969 (2015).
Ropers, F. et al. Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. Hum. Mol. Genet. 20, 2585–2590 (2011).
Elliott, A. M. et al. A novel mutation in KIAA0196: identification of a gene involved in Ritscher-Schinzel/3C syndrome in a First Nations cohort. J. Med. Genet. 50, 819–822 (2013).
Liu, Y. et al. Deficiency of Trim27 protects dopaminergic neurons from apoptosis in the neurotoxin model of Parkinson’s disease. Brain Res. 1588, 17–24 (2014).
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R. & Chan, A. P. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688 (2012).
St Pourcain, B. et al. Common variation contributes to the genetic architecture of social communication traits. Mol. Autism 4, 34 (2013).
Zaman, M. M. et al. Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Mol. Cell. Biol. 33, 4971–4984 (2013).
Inoue, D., Nishimura, K., Kozuka-Hata, H., Oyama, M. & Kitamura, T. The stability of epigenetic factor ASXL1 is regulated through ubiquitination and USP7-mediated deubiquitination. Leukemia 29, 2257–2260 (2015).
Hoischen, A. et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 43, 729–731 (2011).
Magini, P. et al. Two novel patients with Bohring-Opitz syndrome caused by de novo ASXL1 mutations. Am. J. Med. Genet. A 158A, 917–921 (2012).
Russell, B. et al. Clinical management of patients with ASXL1 mutations and Bohring-Opitz syndrome, emphasizing the need for Wilms tumor surveillance. Am. J. Med. Genet. A 167A, 2122–2131 (2015).
Sanz-Pamplona, R. et al. Exome Sequencing Reveals AMER1 as a Frequently Mutated Gene in Colorectal Cancer. Clin. Cancer Res. 21, 4709–4718 (2015).
Untergasser, A. et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Acknowledgements
The authors thank the patients and their families for their wholehearted collaboration. They are also grateful to M. Cozar for the technical assistance, and to the CIBERER Biobank (Valencia, Spain) for handling of the samples, and to CNAG for exome sequencing within the “300 exomes to elucidate rare diseases” program. The authors have no conflict of interest to declare. Funding was from Associació Síndrome Opitz C, Terrassa, Spain; Spanish Ministerio de Economía y Competitividad (SAF2014-56562-R; FECYT, crowdfunding PRECIPITA); Catalan Government (2014SGR932) and from CIBERER (U720). We acknowledge support of the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’.
Author information
Authors and Affiliations
Contributions
R.U., H.F., S.M. and N.R.A. have analysed and validated the Whole Exome Sequencing data, phased the MAGEL2 mutation and manually sequenced the MAGEL2 gene. J.P., L.C., C.C., M.B., S.O., M.M. and J.H. have generated and analysed the Whole Genome sequencing data. A.M.C.G., E.T., J.M.O. and G.N. have clinically evaluated the patients and have generated and written the clinical data. R.U., S.B. and D.G. wrote the main manuscript text. R.U. prepared Figure 1, R.U. and A.M.C.G. and E.T. prepared Table 1, H.F. prepared Table S1, R.U. prepared Table S2, R.U. and S.B. prepared Table S3. B.C. and L.L.V. critically reviewed the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Urreizti, R., Cueto-Gonzalez, A., Franco-Valls, H. et al. A De Novo Nonsense Mutation in MAGEL2 in a Patient Initially Diagnosed as Opitz-C: Similarities Between Schaaf-Yang and Opitz-C Syndromes. Sci Rep 7, 44138 (2017). https://doi.org/10.1038/srep44138
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44138
This article is cited by
-
DPH1 syndrome: two novel variants and structural and functional analyses of seven missense variants identified in syndromic patients
European Journal of Human Genetics (2020)
-
Five new cases of syndromic intellectual disability due to KAT6A mutations: widening the molecular and clinical spectrum
Orphanet Journal of Rare Diseases (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.